1. Introduction
Lipids play an especially important role in plant cells. They not only function as barriers, but also take part in signal transduction, trafficking, morphological changes, and cell division via lipid-protein interactions. To date, many lipid-binding proteins have been identified in plants, including lipid transfer proteins (LTPs). LTPs are small, cationic proteins that are found in all land plants, encoded by large gene families, and expressed in most tissues [
1,
2]. Their abundance indicates a particular importance in plant life.
In LTPs, four conserved disulfide bridges stabilize compact spatial structures containing three or four α-helices and a central hydrophobic cavity suitable for binding varied hydrophobic ligands [
3]. Most often, LTPs are not involved in high-affinity ligand interactions, as evidenced by the micromolar range of dissociation constants (Kd) of LTP-ligand complexes. In vitro experiments showed that a broad range of LTPs bound “standard” hydrophobic ligands such as phospho- and glycolipids, acyl-coenzyme A, fatty acids (FAs) (C12−C27), and prostaglandins [
4]. The preferential ligands are, in most cases, 14−18 carbon FA chains. However, LTP’s in vivo ligand-binding repertoire is still rather under-investigated. To our knowledge, only the non-covalent complex of the peach Pru p 3 with a derivative of the alkaloid camptothecin bound to phytosphingosine [
5], and the covalent complex of the barley LTP with oxylipin [
6], have been isolated from natural sources.
Plant LTPs are mainly synthesized as precursors of a signal peptide, and have extracellular localization. However, some of them can change their localization for an intracellular one, or function as GPI-anchored in membrane proteins [
1]. LTPs are involved in a variety of plant processes due to their ability to bind and transfer different ligands. LTPs transport various hydrophobic molecules inside the cell, in the apoplast and intercellular space as well as through the phloem. These proteins take part in lipid metabolism, the formation of hydrophobic cover layers and protective secondary metabolite secretion. A number of data also indicate that LTPs apparently interact with signaling molecules, for example, with hormones such as jasmonic and gibberellic acids, and play an important role in signal transduction, intercellular interactions and activation of plant immune response [
4]. However, much remains unclear. In this respect, identification of new possible LTP ligands as well as study of protein-ligand interactions may help provide molecular insight into the biological roles of these proteins in plants that are not clearly understood.
In a previous study we showed that Lc-LTP2 from the lentil
Lens culinaris seeds binds and transfers a broad range of FAs (C12-C22) and lysolipids (PC and PG with different acyl chain lengths) [
7]. We described the mechanism of lipid uptake from the LPPG (lyso-palmitoyl phosphatidylglycerol) micelle by Lc-LTP2, and demonstrated the roles of two residues belonging to the “bottom” entrance of Lc-LTP2, Tyr80 and Arg45, which can affect the microarchitecture of the protein hydrophobic cavity, ligand specificity and effectiveness of the FAs and lysolipid binding [
8]. In this study, we identify a new putative Lc-LTP2 ligand and discuss the matter of the detected protein-lipid interactions.
4. Discussion
The lipid composition of eukaryotic cells comprises sphingolipids, neutral lipids, glycolipids, and phospholipids with unique biophysical properties. Most of these lipids form the basis of biomembranes and take part in fat storage, but some of them are involved in signaling and membrane trafficking. Signal lipids are characterized by low abundance, rapid turnover, and transient increase of their synthesis in response to endogenous stimuli, which leads to the activation of different signaling pathways and triggering of subsequent cellular processes. Polyphosphoinositides (PPIs) and phosphatidic acid (PA) play an important role in plant signal transduction. Unlike animals, only five PPIs (i.e., PI(3)P, PI(4)P, PI(5)P, PI(3,5)P2, and PI(4,5)P2) are present in plants. These signaling lipids can specifically interact with proteins via selective lipid-binding domains that recognize the negatively charged lipid heads [
16]. However, protein-ligand interactions depend not only upon electrostatic forces but also on spatial congruence. In the present work, we showed that PI(4,5)P2, mainly located in plasmatic membranes and regulating cellular functions through multiple mechanisms, is a novel signal ligand of the lipid transfer protein Lc-LTP2 from lentil seeds.
It is known that protein-lipid overlay assays are widely used for ligand identification and the study of lipid–protein interactions. As for plant LTPs, only one published paper has used this technique to demonstrate an ability of AtLtpI-4 from Arabidopsis to bind long-chain FAs [
17]. In our experiments, we used hydrophobic nitrocellulose strips spotted with 15 different biologically significant membrane lipids including PA and PPIs. We showed that the lentil Lc-LTP2 as well as LTPs from other plants could bind PI(4,5)P2 (
Figure 1). At the same time, no interaction with other membrane lipids also having two palmitoyl chains was found. We assumed that hydrophobic tails of the adsorbed lipid molecules are mainly anchored to the membrane, and hydrophilic heads were placed on the exposed side of hydrophobic membrane strips [
18]. Primarily, surface electrostatic interactions of cationic plant LTPs were able to take place with the polar heads of lipids. Since we observed such a strong interaction with only PI(4,5)P2, we hypothesized that the ligand-protein conformational congruence was also a key for specific LTPs-lipid binding.
Investigation of PI(4,5)P2 binding in aqueous solution demonstrated that Lc-LTP2 and LTPs from three other plants poorly displaced TNS from the protein cavities (
Figure 2). We assumed that the cause lies in the conformational features of the formed protein-lipid complexes. On the one hand, incomplete space-filling of the protein hydrophobic cavities with the ligand might also take place. We previously observed such an effect for LMPG and LMPC, which had shorter acyl tails than LPPG and LPPC and did a worse job of displacing TNS [
19]. On the other hand, the LTP-PI(4,5)P2 complexes in protein-overlay assays might be formed not only due to ligand interactions with the hydrophobic cavities, but also with other sites on the protein surfaces.
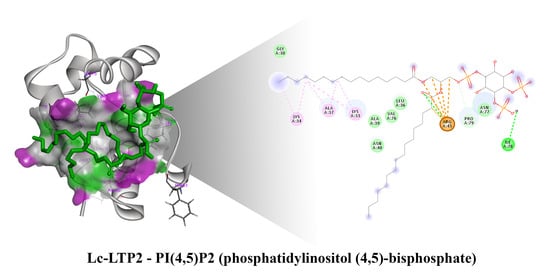
In order to clarify the situation, a computer modeling of the Lc-LTP2 complex was performed with PI(4,5)P2. In previous studies of NMR and molecular docking we showed that Lc-LTP2 formed complexes with anionic and zwitterionic phospholipids (DMPG, DHPC (1,2-diphytanoyl-sn-glycero-3-phosphocholine), LPPG and LPPC) in which the hydrophobic tails of the ligands were located inside the protein cavity, and their heads were located near the “top” entrance of the hydrophobic cavity [
7]. In this work, we used the NMR structures of apo-Lc-LTP2 (PDB ID: 2MAL) for evaluation of an initial step of the lipid contact with the protein and the Lc-LTP2 spatial structure in its complex with LPPG (PDB ID: 5LQV]). This was used to model a possible final ligand location in the protein hydrophobic cavity (
Figure 3). All the 10 calculated conformations showed that the ligand was located on the protein surface close to the “bottom” entrance of apo-Lc-LTP2 (
Figure 3A). We found that Arg45 played a key role in the interaction with the ligand, forming a hydrogen bond and electrostatic interactions with the polar head of PI(4,5)P2 (
Figure 4). We assumed that a similar ligand orientation in the complex with LTP might take place in the case of protein-overlay assays. As for the structure of liganded Lc-LTP2, hydrophobic tails of PI(4,5)P2 were located inside the protein cavity in all 10 calculated conformations, but its polar head was located outside the cavity close to the “bottom” entrance of the cavity. Thus, orientation of PI(4,5)P2 inside the Lc-LTP2 hydrophobic cavity near the “bottom” entrance was different compared with the abovementioned phospholipids, with one or two acyl chains accommodating at an opposite entrance. Based on these data, it can be concluded that Arg45 plays a significant role both in initiating surface interactions and holding the ligand in the hydrophobic cavity. Further, this might be applicable not only to Arg45, but also to Tyr80, Lys53 and some other amino acids that play an important role in protein-ligand complex stabilization (
Figure 4).
Earlier, we showed that Lc-LTP2 induced a significant leakage of negatively charged POPG vesicles, but did not affect zwitterionic POPC vesicles or POPC:POPG vesicle permeability even at a ratio of 1:1 of these lipids. It was concluded that electrostatic interactions play a crucial role in the accumulation and destabilization of the protein vesicle surface [
7]. Here, we included PI(4,5)P2 as a minor component (2%) of cell membranes in vesicle compositions. It was demonstrated that an inclusion of this lipid did not affect the permeabilizing activity of Lc-LTP2 in the case of POPG vesicles, but it had a slight effect on POPC vesicles (
Figure 5). We assumed that the inclusion of PI(4,5)P2 does not affect the efficiency of POPG:PI(4,5)P2 (98:2) vesicle destruction by Lc-LTP2 because they had the negative charge across their surface, which probably makes the specific interaction of the protein with a polar head of PI(4,5)P2 impossible. Since POPC:PI(4,5)P2 (98:2) liposomes are almost uncharged, such a weak effect, in our opinion, could be associated not only with the electrostatic interaction of Lc-LTP2 and PI(4,5)P2, but mostly with the conformational congruence of this ligand to the protein.
In our previous work, we obtained three mutant analogs of Lc-LTP2: R45A, Y80A and R45A/Y80A, and showed an important role of Arg45 and Tyr80 residues located at the “bottom” entrance of the protein cavity in the binding of both FAs and lysolipids [
8]. Here, we used these mutant analogs to study a role of Arg45 and Tyr80 residues in Lc-LTP2 interactions with PI(4,5)P2. We demonstrated that all the three mutant analogs bound this ligand worse than Lc-LTP2, especially R45A (
Figure 2). These data confirm a significant role of Arg45 in PI(4,5)P2 binding as the residue that gave shape to the cavity architectonics. Tyr80 is probably also involved in the formation of a complex with the tested ligand, but to a lesser extent than Arg45. It is interesting to note that R45A/Y80A has the largest hydrophobic cavity among all the mutant analogs [
8], and binds PI(4,5)P2 slightly better than other mutant analogs, probably due to a deeper placement in the protein cavity.
Molecular docking data showed that PI(4,5)P2 interacted with the “bottom” entrance of the wild-type protein and all of the mutant analogs of apo-Lc-LTP2. At the same time, an increase in the cavity volume (R45A < Y80A < R45A/Y80A) led to a deeper placement of the ligand in the cavity, which confirmed the results of the binding experiments (
Figure 3). It is worth noting that PI(4,5)P2 orientation in the structures of all liganded mutant analogs was the same as that of Lc-LTP2. We assumed that PI(4,5)P2 has a very bulky polar head, which makes it impossible to place this ligand into the protein cavity in B and C orientations, as was demonstrated for LPPC, LPPG, DMPG and DHPC [
7,
8]. We previously described the mechanism of lipid uptake according to which Lc-LTP2 binds to the surface of LPPG micelle by its positively charged “bottom” entrance located near the C-terminal tail of the protein, after which the lipid penetrates into the protein cavity [
7]. Based on our current data, we assume that in the case of signal function of PI(4,5)P2 such a mechanism of lipid uptake is less favorable upon contact with the membrane.
In experiments with vesicles of different compositions we unexpectedly found that displacement of Arg45 did not affect the permeabilizing potential of Lc-LTP2 in the case of POPG. At the same time, the R45A/Y80A and Y80A mutant analogs had no effects on vesicle permeability at all. Thus, the absence of Arg45 presumably took part in an initial electrostatic interaction in which the vesicles could be compensated by the adjacently charged Lys81 and Lys53, which are also located close to the “bottom” entrance of Lc-LTP2. In contrast, the obtained data proved that the absence of Tyr80 cannot be compensated by any neighboring amino acids. At this point, we presume that Tyr80 is probably the key amino acid residue playing an essential role in Lc-LTP2-membrane docking.
5. Conclusions
In the present work, we showed that PI(4,5)P2 is a new possible ligand of lentil Lc-LTP2 and other plant LTPs using protein-lipid overlay assays. Results of PI(4,5)P2 binding in solution, molecular docking and liposome leakage experiments allowed us to assume that Lc-LTP2 binds this lipid in a different way than in cases of phosphatidylcholines and phosphatidylglycerols with one or two acyl tails. We showed that in the solution Lc-LTP2 apparently formed a complex with PI(4,5)P2 in which a polar head of the ligand is located outside the protein cavity close to the “bottom” entrance. At the same time, in the case of PI(4,5)P2 adsorbed on the membrane strip along with the electrostatic surface interaction of the protein with the lipid polar head, the conformational congruence of Lc-LTP2 with the ligan might be the most important factor behind the complex formation. We also demonstrated that Arg45, conserved in most plant LTPs, plays an important role in PI(4,5)P2-protein binding. Here, we suggested that Lc-LTP2 might bind to the polar head of PI(4,5)P2 without capturing the lipid from the cell membrane, after which a cascade of some signaling reactions could be triggered or other events might take place. Moreover, in addition to our previous data on the importance of another conservative residue, Tyr80, in the stabilization of the LTP-lipid complexes, we showed here for the first time a key role of this residue in LTP-membrane docking.