A Glimpse into the Diverse Cellular Immunity against SARS-CoV-2
Abstract
:1. Importance
2. Introduction
3. Materials and Methods
3.1. Blood Donors and PBMC Isolation
3.2. Synthesis of Overlapping Pentadecamer Peptides
3.3. In Vitro Generation of SARS-CoV-2-Specific Cytotoxic T Lymphocytes
3.4. ELISPOT Assay of Immune Effector Function
3.5. Immune Effector CD107a Degranulation and Intracellular Cytokine Staining
3.6. Statistical Analysis
4. Results
4.1. The Rationale for the Design of Antigenic Peptide Pools of SARS-CoV-2 Structural and Functional Proteins
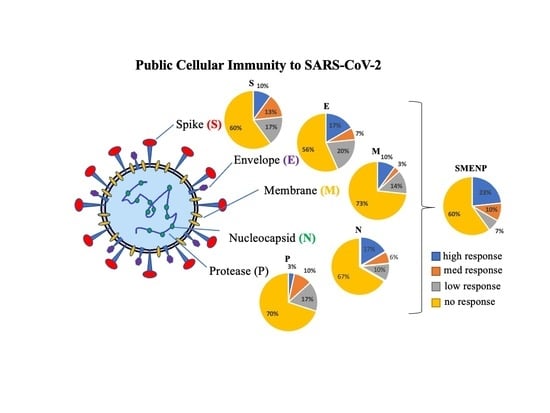
4.2. Assessment of SARS-CoV-2 Antigen-Specific Precursor Frequency in Healthy Individuals
4.3. Activation of SARS-CoV-2-Specific Immune Cells In Vitro
4.4. Immune Re-Stimulation and Extended Culture to Enhance Detection of Low-Frequency Viral Antigen-Specific Responding Cells
4.5. Anti-SARS-CoV-2 Effector Activities of the In Vitro DC-SEMNP-Activated T Cells
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological Findings of COVID-19 Associated with Acute Respiratory Distress Syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Coperchini, F.; Chiovato, L.; Croce, L.; Magri, F.; Rotondi, M. The Cytokine Storm in COVID-19: An Overview of the Involvement of the Chemokine/Chemokine-Receptor System. Cytokine Growth Factor Rev. 2020, 53, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.-W.; Yuan, S.; Kok, K.-H.; To, K.K.-W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.; Poon, R.W.-S.; et al. A Familial Cluster of Pneumonia Associated with the 2019 Novel Coronavirus Indicating Person-to-Person Transmission: A Study of a Family Cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef] [Green Version]
- Birra, D.; Benucci, M.; Landolfi, L.; Merchionda, A.; Loi, G.; Amato, P.; Licata, G.; Quartuccio, L.; Triggiani, M.; Moscato, P. COVID 19: A Clue from Innate Immunity. Immunol. Res. 2020, 68, 161–168. [Google Scholar] [CrossRef]
- Tan, M.; Liu, Y.; Zhou, R.; Deng, X.; Li, F.; Liang, K.; Shi, Y. Immunopathological Characteristics of Coronavirus Disease 2019 Cases in Guangzhou, China. Immunology 2020, 160, 261–268. [Google Scholar] [CrossRef]
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020, 71, 762–768. [Google Scholar] [CrossRef]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and Immunological Features of Severe and Moderate Coronavirus Disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef] [Green Version]
- Wan, Y.; Shang, J.; Sun, S.; Tai, W.; Chen, J.; Geng, Q.; He, L.; Chen, Y.; Wu, J.; Shi, Z.; et al. Molecular Mechanism for Antibody-Dependent Enhancement of Coronavirus Entry. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwasaki, A.; Yang, Y. The Potential Danger of Suboptimal Antibody Responses in COVID-19. Nat. Rev. Immunol. 2020, 20, 339–341. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Wang, L.; Kong, X.; Geng, J.; Xiao, D.; Ma, C.; Jiang, X.-M.; Wang, P.-H. Long-Term Coexistence of SARS-CoV-2 with Antibody Response in COVID-19 Patients. J. Med. Virol. 2020, 92, 1684–1689. [Google Scholar] [CrossRef]
- Casadevall, A.; Pirofski, L.-A. The Convalescent Sera Option for Containing COVID-19. J. Clin. Investig. 2020, 130, 1545–1548. [Google Scholar] [CrossRef] [Green Version]
- Odak, I.; Barros-Martins, J.; Bošnjak, B.; Stahl, K.; David, S.; Wiesner, O.; Busch, M.; Hoeper, M.M.; Pink, I.; Welte, T.; et al. Reappearance of Effector T Cells Is Associated with Recovery from COVID-19. EBioMedicine 2020, 57, 102885. [Google Scholar] [CrossRef]
- Altmann, D.M.; Boyton, R.J. SARS-CoV-2 T Cell Immunity: Specificity, Function, Durability, and Role in Protection. Sci. Immunol. 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Gao, Z.-Y.; Chang, L.-J.; Liang, Y.; Tan, X.-Y.; Liu, J.-H.; Yu, X.-J.; Yang, F.-H.; Xie, Y.; Lu, D.-P. Adoptive Transfer of Cytomegalovirus/Epstein-Barr Virus-Specific Immune Effector Cells for Therapeutic and Preventive/Preemptive Treatment of Pediatric Allogeneic Cell Transplant Recipients. J. Pediatr. Hematol. Oncol. 2010, 32, e31–e37. [Google Scholar] [CrossRef]
- Han, S.; Huang, Y.; Liang, Y.; Ho, Y.; Wang, Y.; Chang, L.-J. Phenotype and Functional Evaluation of Ex Vivo Generated Antigen-Specific Immune Effector Cells with Potential for Therapeutic Applications. J. Hematol. Oncol. 2009, 2, 34. [Google Scholar] [CrossRef] [Green Version]
- Rooney, C.M.; Smith, C.A.; Ng, C.Y.; Loftin, S.K.; Sixbey, J.W.; Gan, Y.; Srivastava, D.K.; Bowman, L.C.; Krance, R.A.; Brenner, M.K.; et al. Infusion of Cytotoxic T Cells for the Prevention and Treatment of Epstein-Barr Virus-Induced Lymphoma in Allogeneic Transplant Recipients. Blood 1998, 92, 1549–1555. [Google Scholar] [CrossRef]
- Betts, M.R.; Brenchley, J.M.; Price, D.A.; De Rosa, S.C.; Douek, D.C.; Roederer, M.; Koup, R.A. Sensitive and Viable Identification of Antigen-Specific CD8+ T Cells by a Flow Cytometric Assay for Degranulation. J. Immunol. Methods 2003, 281, 65–78. [Google Scholar] [CrossRef]
- Chen, X.; He, J.; Chang, L.-J. Alteration of T Cell Immunity by Lentiviral Transduction of Human Monocyte-Derived Dendritic Cells. Retrovirology 2004, 1, 37. [Google Scholar] [CrossRef] [Green Version]
- Peters, P.J.; Borst, J.; Oorschot, V.; Fukuda, M.; Krähenbühl, O.; Tschopp, J.; Slot, J.W.; Geuze, H.J. Cytotoxic T Lymphocyte Granules Are Secretory Lysosomes, Containing Both Perforin and Granzymes. J. Exp. Med. 1991, 173, 1099–1109. [Google Scholar] [CrossRef]
- Kannan, K.; Stewart, R.M.; Bounds, W.; Carlsson, S.R.; Fukuda, M.; Betzing, K.W.; Holcombe, R.F. Lysosome-Associated Membrane Proteins h-LAMP1 (CD107a) and h-LAMP2 (CD107b) Are Activation-Dependent Cell Surface Glycoproteins in Human Peripheral Blood Mononuclear Cells Which Mediate Cell Adhesion to Vascular Endothelium. Cell Immunol. 1996, 171, 10–19. [Google Scholar] [CrossRef]
- Gao, Q.; Bao, L.; Mao, H.; Wang, L.; Xu, K.; Yang, M.; Li, Y.; Zhu, L.; Wang, N.; Lv, Z.; et al. Development of an Inactivated Vaccine Candidate for SARS-CoV-2. Science 2020, 369, 77–81. [Google Scholar] [CrossRef]
- Corey, L.; Mascola, J.R.; Fauci, A.S.; Collins, F.S. A Strategic Approach to COVID-19 Vaccine R&D. Science 2020, 368, 948–950. [Google Scholar] [CrossRef]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-Specific T Cell Immunity in Cases of COVID-19 and SARS, and Uninfected Controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef]
- Tang, F.; Quan, Y.; Xin, Z.-T.; Wrammert, J.; Ma, M.-J.; Lv, H.; Wang, T.-B.; Yang, H.; Richardus, J.H.; Liu, W.; et al. Lack of Peripheral Memory B Cell Responses in Recovered Patients with Severe Acute Respiratory Syndrome: A Six-Year Follow-up Study. J. Immunol. 2011, 186, 7264–7268. [Google Scholar] [CrossRef] [Green Version]
- Sekine, T.; Perez-Potti, A.; Rivera-Ballesteros, O.; Strålin, K.; Gorin, J.-B.; Olsson, A.; Llewellyn-Lacey, S.; Kamal, H.; Bogdanovic, G.; Muschiol, S.; et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell 2020, 183, 158–168.e14. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501. [Google Scholar] [CrossRef]
- Mateus, J.; Grifoni, A.; Tarke, A.; Sidney, J.; Ramirez, S.I.; Dan, J.M.; Burger, Z.C.; Rawlings, S.A.; Smith, D.M.; Phillips, E.; et al. Selective and Cross-Reactive SARS-CoV-2 T Cell Epitopes in Unexposed Humans. Science 2020, 370, 89–94. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Neumann-Haefelin, C. HLA-B27-Mediated Protection in HIV and Hepatitis C Virus Infection and Pathogenesis in Spondyloarthritis: Two Sides of the Same Coin? Curr. Opin. Rheumatol. 2013, 25, 426–433. [Google Scholar] [CrossRef]
- Forster, P.; Forster, L.; Renfrew, C.; Forster, M. Phylogenetic Network Analysis of SARS-CoV-2 Genomes. Proc. Natl. Acad. Sci. USA 2020, 117, 9241–9243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanley, P.J.; Shaffer, D.R.; Cruz, C.R.Y.; Ku, S.; Tzou, B.; Liu, H.; Demmler-Harrison, G.; Heslop, H.E.; Rooney, C.M.; Gottschalk, S.; et al. Expansion of T Cells Targeting Multiple Antigens of Cytomegalovirus, Epstein-Barr Virus and Adenovirus to Provide Broad Antiviral Specificity after Stem Cell Transplantation. Cytotherapy 2011, 13, 976–986. [Google Scholar] [CrossRef] [Green Version]
- Alshukairi, A.N.; Khalid, I.; Ahmed, W.A.; Dada, A.M.; Bayumi, D.T.; Malic, L.S.; Althawadi, S.; Ignacio, K.; Alsalmi, H.S.; Al-Abdely, H.M.; et al. Antibody Response and Disease Severity in Healthcare Worker MERS Survivors. Emerg. Infect. Dis. 2016, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, H.-S.; Kim, Y.; Kim, G.; Lee, J.Y.; Jeong, I.; Joh, J.-S.; Kim, H.; Chang, E.; Sim, S.Y.; Park, J.-S.; et al. Immune Responses to Middle East Respiratory Syndrome Coronavirus During the Acute and Convalescent Phases of Human Infection. Clin. Infect. Dis. 2019, 68, 984–992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibarrondo, F.J.; Fulcher, J.A.; Goodman-Meza, D.; Elliott, J.; Hofmann, C.; Hausner, M.A.; Ferbas, K.G.; Tobin, N.H.; Aldrovandi, G.M.; Yang, O.O. Rapid Decay of Anti-SARS-CoV-2 Antibodies in Persons with Mild Covid-19. N. Engl. J. Med. 2020, 383, 1085–1087. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, C.-W.; Liu, Y.; Jiao, C.; Liu, H.; Gong, J.; Chen, X.; Chang, L.-J. A Glimpse into the Diverse Cellular Immunity against SARS-CoV-2. Vaccines 2021, 9, 827. https://doi.org/10.3390/vaccines9080827
Chang C-W, Liu Y, Jiao C, Liu H, Gong J, Chen X, Chang L-J. A Glimpse into the Diverse Cellular Immunity against SARS-CoV-2. Vaccines. 2021; 9(8):827. https://doi.org/10.3390/vaccines9080827
Chicago/Turabian StyleChang, Cheng-Wei, Yuchen Liu, Cheng Jiao, Hongwei Liu, Jie Gong, Xiaochuan Chen, and Lung-Ji Chang. 2021. "A Glimpse into the Diverse Cellular Immunity against SARS-CoV-2" Vaccines 9, no. 8: 827. https://doi.org/10.3390/vaccines9080827