NGFR Gene and Single Nucleotide Polymorphisms, rs2072446 and rs11466162, Playing Roles in Psychiatric Disorders
Abstract
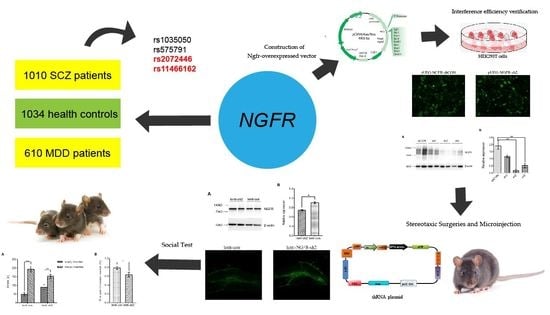
:1. Introduction
2. Materials and Methods
2.1. Participants and Animals
2.2. DNA Extraction and Genotyping
2.3. Statistical Analysis
2.4. Behavioral Tests on Ngfr-Knock-Down Mice
2.4.1. Construction of Ngfr-Interference Plasmid
2.4.2. Construction of Ngfr-Overexpressed Vector
2.4.3. Interference Efficiency Verification
2.4.4. Stereotaxic Surgeries and Microinjection
2.4.5. Behavioral Tests
3. Results
3.1. Association-Study Analysis of the NGFR SNPs with Schizophrenia
3.2. Correlation between NGFR Gene Polymorphisms and MDD
3.3. Ngfr-Knockdown Mice Showing a Trend of Social Avoidance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Hallmayer, J.; Cleveland, S.; Torres, A.; Phillips, J.; Cohen, B.; Torigoe, T.; Miller, J.; Fedele, A.; Collins, J.; Smith, K. Genetic heritability and shared environmental factors among twin pairs with autism. Arch. Gen. Psychiatry 2011, 68, 1095–1102. [Google Scholar] [CrossRef]
- Tsuang, M.T.; Bar, J.L.; Stone, W.S.; Faraone, S.V. Gene-environment interactions in mental disorders. World Psychiatry 2004, 3, 73. [Google Scholar] [PubMed]
- Shih, R.A.; Belmonte, P.L.; Zandi, P.P. A review of the evidence from family, twin and adoption studies for a genetic contribution to adult psychiatric disorders. Int. Rev. Psychiatry 2004, 16, 260–283. [Google Scholar] [CrossRef] [PubMed]
- Nyatega, C.O.; Qiang, L.; Adamu, M.J.; Younis, A.; Kawuwa, H.B. Altered Dynamic Functional Connectivity of Cuneus in Schizophrenia Patients: A Resting-State fMRI Study. Appl. Sci. 2021, 11, 11392. [Google Scholar] [CrossRef]
- Rog, J.; Błażewicz, A.; Juchnowicz, D.; Ludwiczuk, A.; Stelmach, E.; Kozioł, M.; Karakula, M.; Niziński, P.; Karakula-Juchnowicz, H. The Role of GPR120 Receptor in Essential Fatty Acids Metabolism in Schizophrenia. Biomedicines 2020, 8, 243. [Google Scholar] [CrossRef]
- Silveira, S.; Hecht, M.; Adli, M.; Voelkle, M.C.; Singer, T. Exploring the Structure and Interrelations of Time-Stable Psychological Resilience, Psychological Vulnerability, and Social Cohesion. Front. Psychiatry 2022, 13, 804763. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Szabó, Á.; Spekker, E.; Polyák, H.; Tóth, F.; Vécsei, L. Mitochondrial Impairment: A Common Motif in Neuropsychiatric Presentation? The Link to the Tryptophan—Kynurenine Metabolic System. Cells 2022, 11, 2607. [Google Scholar] [CrossRef]
- Gaebler, A.J.; Finner-Prével, M.; Sudar, F.P.; Langer, F.H.; Keskin, F.; Gebel, A.; Zweerings, J.; Mathiak, K. The Interplay between Vitamin D, Exposure of Anticholinergic Antipsychotics and Cognition in Schizophrenia. Biomedicines 2022, 10, 1096. [Google Scholar] [CrossRef]
- Tanaka, M.; Spekker, E.; Szabó, Á.; Polyák, H.; Vécsei, L. Modelling the neurodevelopmental pathogenesis in neuropsychiatric disorders. Bioactive kynurenines and their analogues as neuroprotective agents—In celebration of 80th birthday of Professor Peter Riederer. J. Neural Transm. 2022, 129, 627–642. [Google Scholar] [CrossRef]
- Avan, R.; Sahebnasagh, A.; Hashemi, J.; Monajati, M.; Faramarzi, F.; Henney, N.C.; Montecucco, F.; Jamialahmadi, T.; Sahebkar, A. Update on Statin Treatment in Patients with Neuropsychiatric Disorders. Life 2021, 11, 1365. [Google Scholar] [CrossRef] [PubMed]
- Wiescholleck, V.; Manahan-Vaughan, D. Persistent deficits in hippocampal synaptic plasticity accompany losses of hippocampus-dependent memory in a rodent model of psychosis. Front. Integr. Neurosci. 2013, 7, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McAllister, A.K.; Katz, L.C.; Lo, D.C. Neurotrophins and synaptic plasticity. Annu. Rev. Neurosci. 1999, 22, 295–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraemer, B.R.; Yoon, S.O.; Carter, B.D. The Biological Functions and Signaling Mechanisms of the p75 Neurotrophin Receptor. Handb. Exp. Pharmacol. 2014, 220, 121–164. [Google Scholar] [PubMed]
- Zhou, X.F.; Rush, R.A.; McLachlan, E.M. Differential expression of the p75 nerve growth factor receptor in glia and neurons of the rat dorsal root ganglia after peripheral nerve transection. J. Neurosci. 1996, 16, 2901–2911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- VanGuilder Starkey, H.D.; Sonntag, W.E.; Freeman, W.M. Increased hippocampal NgR1 signaling machinery in aged rats with deficits of spatial cognition. Eur. J. Neurosci. 2013, 37, 1643–1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malik, S.C.; Sozmen, E.G.; Baeza-Raja, B.; Le Moan, N.; Akassoglou, K.; Schachtrup, C. In vivo functions of p75(NTR): Challenges and opportunities for an emerging therapeutic target. Trends Pharmacol. Sci. 2021, 42, 772–788. [Google Scholar] [CrossRef]
- Zhou, L.; Xiong, J.; Lim, Y.; Ruan, Y.; Huang, C.; Zhu, Y.; Zhong, J.H.; Xiao, Z.; Zhou, X.F. Upregulation of blood proBDNF and its receptors in major depression. J. Affect. Disord. 2013, 150, 776–784. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Chen, S.; Li, C.; Lu, N.; Yue, Y.; Yin, Y.; Zhang, Y.; Zhi, X.; Zhang, D.; Yuan, Y. The serum protein levels of the tPA-BDNF pathway are implicated in depression and antidepressant treatment. Transl. Psychiatry 2017, 7, e1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zakharyan, R.; Atshemyan, S.; Gevorgyan, A.; Boyajyan, A. Nerve growth factor and its receptor in schizophrenia. BBA Clin. 2014, 1, 24–29. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Jiang, H.; Liu, Y.; Hou, Z.; Yue, Y.; Zhang, Y.; Zhao, F.; Xu, Z.; Li, Y.; Mou, X.; et al. Combined serum levels of multiple proteins in tPA-BDNF pathway may aid the diagnosis of five mental disorders. Sci. Rep. 2017, 7, 6871. [Google Scholar] [CrossRef] [PubMed]
- Kunugi, H.; Hashimoto, R.; Yoshida, M.; Tatsumi, M.; Kamijima, K. A missense polymorphism (S205L) of the low-affinity neurotrophin receptor p75NTR gene is associated with depressive disorder and attempted suicide. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2004, 129, 44–46. [Google Scholar] [CrossRef] [PubMed]
- Gau, Y.T.; Liou, Y.J.; Yu, Y.W.; Chen, T.J.; Lin, M.W.; Tsai, S.J.; Hong, C.J. Evidence for association between genetic variants of p75 neurotrophin receptor (p75NTR) gene and antidepressant treatment response in Chinese major depressive disorder. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2008, 147, 594–599. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.Y.; Ruan, C.S.; Yang, C.R.; Li, J.Y.; Kang, Z.L.; Zhou, L.; Liu, D.; Zeng, Y.Q.; Wang, T.H.; Tian, C.F.; et al. ProBDNF Signaling Regulates Depression-Like Behaviors in Rodents under Chronic Stress. Neuropsychopharmacology 2016, 41, 2882–2892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, L.; Zhou, X.F.; Bobrovskaya, L. Blockage of p75 NTR ameliorates depressive-like behaviours of mice under chronic unpredictable mild stress. Behav. Brain Res. 2020, 396, 112905. [Google Scholar] [CrossRef] [PubMed]
- Catts, V.S.; Al-Menhali, N.; Burne, T.H.J.; Colditz, M.J.; Coulson, E.J. The p75 neurotrophin receptor regulates hippocampal neurogenesis and related behaviours. Eur. J. Neurosci. 2008, 28, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Olsen, D.; Kaas, M.; Schwartz, O.; Nykjaer, A.; Glerup, S. Loss of BDNF or its receptors in three mouse models has unpredictable consequences for anxiety and fear acquisition. Learn. Mem. 2013, 20, 499–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.-F.; Li, E.; Huber, L.J.; Landis, S.C.; Sharpe, A.H.; Chao, M.V.; Jaenisch, R. Targeted mutation of the gene encoding the low affinity NGF receptor p75 leads to deficits in the peripheral sensory nervous system. Cell 1992, 69, 737–749. [Google Scholar] [CrossRef]
- Puschban, Z.; Sah, A.; Grutsch, I.; Singewald, N.; Dechant, G. Reduced Anxiety-Like Behavior and Altered Hippocampal Morphology in Female p75NTR(exon IV−/−) Mice. Front. Behav. Neurosci. 2016, 10, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinowich, K.; Schloesser, R.J.; Lu, Y.; Jimenez, D.V.; Paredes, D.; Greene, J.S.; Greig, N.H.; Manji, H.K.; Lu, B. Roles of p75(NTR), long-term depression, and cholinergic transmission in anxiety and acute stress coping. Biol. Psychiatry 2012, 71, 75–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, X.L.; Zou, J.X.; Wu, R.Y.; Yang, Y.; Tai, F.D.; Zeng, S.Y.; Jia, R.; Zhang, X.; Liu, E.Q.; Broders, H. Strain and sex differences in anxiety-like and social behaviors in C57BL/6J and BALB/cJ mice. Exp. Anim. 2011, 60, 111–123. [Google Scholar] [CrossRef] [PubMed]





| SNP ID a | Chromosome b | Location | Polymorphisms c | |
|---|---|---|---|---|
| NGFR | rs1035050 | 17:49486650 | promoter | T/C |
| rs575791 | 17:49497393 | intron | A/G | |
| rs2072446 | 17:49510457 | missense | C/T | |
| rs11466162 | 17:49513533 | 3 Prime UTR Variant | G/A |
| Ngfr-sh1-F. | GATCCCGGGCCTTGTGGCCTATATTCTCAAGAGAAATATAGGCCACAAGGCCCTTTTTT |
| Ngfr-sh1-R | CTAGAAAAAAGGGCCTTGTGGCCTATATTTCTCTTGAGAATATAGGCCACAAGGCCCGG |
| Ngfr-sh2-F | GATCCCGGTCGAGAAGCTGCTCAATTTCAAGAGAATTGAGCAGCTTCTCGACCTTTTTT |
| Ngfr-sh2-R | CTAGAAAAAAGGTCGAGAAGCTGCTCAATTCTCTTGAAATTGAGCAGCTTCTCGACCGG |
| Ngfr-sh3-F | GATCCCGCATCCAGAGAGCTGACATTTCAAGAGAATGTCAGCTCTCTGGATGCTTTTTT |
| Ngfr-sh3-R | CTAGAAAAAAGCATCCAGAGAGCTGACATTCTCTTGAAATGTCAGCTCTCTGGATGCGG |
| shCON-F | GATCCCGTTCTCCGAACGTGTCACGTTTCAAGAGATGCACTGTGCAAGCCTCTTTTTT |
| shCON-R | CTAGAAAAAAGAGGCTTGCACAGTGCATCTCTTGAAACGTGACACGTTCGGAGAACGG |
| SNP | Genotype Frequency | χ2 | p Value a | Allele Frequency | χ2 | p Value a | Odds Ratio (95%CI) | HWE p Value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | C | T | ||||||||
| rs1035050 | SCZ | 570 (0.576) | 362 (0.366) | 57 (0.058) | 1.01 | 0.605 | 1502 (0.759) | 476 (0.241) | 0.99 | 0.320 | 0.929 (0.803–1.07) | 0.96 |
| Control | 613 (0.596) | 364 (0.354) | 52 (0.051) | 1590 (0.773) | 468 (0.227) | 0.83 | ||||||
| AA | AG | GG | A | G | ||||||||
| rs575791 | SCZ | 609 (0.604) | 352 (0.349) | 48 (0.048) | 1.46 | 0.482 | 1570 (0.778) | 448 (0.222) | 1.40 | 0.237 | 0.914 (0.787–1.06) | 0.75 |
| Control | 645 (0.626) | 344 (0.334) | 41 (0.040) | 1634 (0.793) | 426 (0.207) | 0.56 | ||||||
| CC | CT | TT | C | T | ||||||||
| rs2072446 | SCZ | 808 (0.813) | 169 (0.170) | 17 (0.017) | 8.23 | 0.016 | 1785 (0.898) | 203 (0.102) | 4.17 | 0.041 | 1.23 (1.01–1.49) | 0.022 |
| Control | 784 (0.768) | 224 (0.219) | 13 (0.013) | 1792 (0.878) | 250 (0.122) | 0.50 | ||||||
| AA | AG | GG | A | G | ||||||||
| rs11466162 | SCZ | 13 (0.013) | 143 (0.142) | 853 (0.845) | 11.50 | 0.0032 | 169 (0.084) | 1849 (0.916) | 3.92 | 0.048 | 0.807 (0.652–0.998) | 0.015 |
| Control | 6 (0.006) | 197 (0.192) | 824 (0.802) | 209 (0.102) | 1845 (0.898) | 0.11 | ||||||
| Scheme 2 | Genotype Frequency | χ2 | p Value a | Allele Frequency | χ2 | p Value a | Odds Ratio (95%CI) | H-W p Value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | C | T | ||||||||
| rs1035050 | MDD | 361 (0.605) | 201 (0.337) | 35 (0.059) | 0.83 | 0.661 | 923 (0.773) | 271 (0.227) | 0.00082 | 0.977 | 1.00 (0.846–1.19) | 0.32 |
| Control | 613 (0.596) | 364 (0.354) | 52 (0.051) | 1590 (0.773) | 468 (0.227) | 0.83 | ||||||
| AA | AG | GG | A | G | ||||||||
| rs575791 | MDD | 349 (0.589) | 216 (0.364) | 28 (0.047) | 2.36 | 0.308 | 914 (0.771) | 272 (0.229) | 2.27 | 0.132 | 0.876 (0.737–1.04) | 0.46 |
| Control | 645 (0.626) | 344 (0.334) | 41 (0.040) | 1634 (0.793) | 426 (0.207) | 0.56 | ||||||
| CC | CT | TT | C | T | ||||||||
| rs2072446 | MDD | 479 (0.800) | 114 (0.190) | 6 (0.010) | 2.26 | 0.324 | 1072 (0.895) | 126 (0.105) | 2.19 | 0.139 | 1.19 (0.946–1.49) | 0.79 |
| Control | 784 (0.768) | 224 (0.219) | 13 (0.013) | 1792 (0.878) | 250 (0.122) | 0.50 | ||||||
| AA | AG | GG | A | G | ||||||||
| rs11466162 | MDD | 7 (0.012) | 107 (0.182) | 475 (0.806) | 1.91 | 0.3844 | 121 (0.103) | 1057 (0.897) | 0.0076 | 0.931 | 1.01 (0.798–1.28) | 0.73 |
| Control | 6 (0.006) | 197 (0.192) | 824 (0.802) | 209 (0.102) | 1845 (0.898) | 0.11 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, L.; Hou, B.; Ji, L.; Ren, D.; Yuan, F.; Liu, L.; Bi, Y.; Yang, F.; Yu, S.; Yi, Z.; et al. NGFR Gene and Single Nucleotide Polymorphisms, rs2072446 and rs11466162, Playing Roles in Psychiatric Disorders. Brain Sci. 2022, 12, 1372. https://doi.org/10.3390/brainsci12101372
Zhao L, Hou B, Ji L, Ren D, Yuan F, Liu L, Bi Y, Yang F, Yu S, Yi Z, et al. NGFR Gene and Single Nucleotide Polymorphisms, rs2072446 and rs11466162, Playing Roles in Psychiatric Disorders. Brain Sciences. 2022; 12(10):1372. https://doi.org/10.3390/brainsci12101372
Chicago/Turabian StyleZhao, Longyou, Binyin Hou, Lei Ji, Decheng Ren, Fan Yuan, Liangjie Liu, Yan Bi, Fengping Yang, Shunying Yu, Zhenghui Yi, and et al. 2022. "NGFR Gene and Single Nucleotide Polymorphisms, rs2072446 and rs11466162, Playing Roles in Psychiatric Disorders" Brain Sciences 12, no. 10: 1372. https://doi.org/10.3390/brainsci12101372