Glioblastoma Multiforme Tumors in Women Have a Lower Expression of Fatty Acid Elongases ELOVL2, ELOVL5, ELOVL6, and ELOVL7 than in Men
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Samples
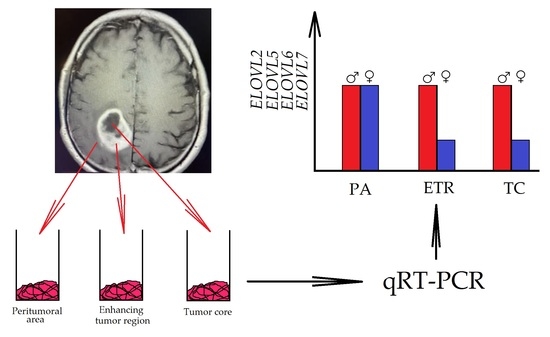
- The peritumoral area, a buffer zone between the GBM tumor and healthy tissue, with individual foci of infiltration. The peritumoral area is considered an appropriate control for the study of GBM tumors [24]. For this reason, the results obtained from GBM tumor studies were assimilated into the peritumoral area.
- The non-enhancing tumor core. This area of GBM is usually located in the central part of the GBM tumor,
- The enhancing tumor region, surrounding the tumor core.
2.2. Cell Cultures and Treatment
2.3. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
- for GAPDH, (5′-TCATGGGTGTGAACCATGAGAA-3′ and 5′-GGCATGGACTGTGGTCATGAG-3′);
- for ELOVL1, (5′-TTATTCTCCGAAAGAAAGACGGG-3′ and 5′-ATGACATGCACGGAAGAGTTTAT-3′)
- for ELOVL2, (5′-ATGTTTGGACCGCGAGATTCT-3′ and 5′-CCCAGCCATATTGAGAGCAGATA-3′);
- for ELOVL3, (5′-CTGTTCCAGCCCTATAACTTCG-3′ and 5′-GAATGAGGTTGCCCAATACTCC-3′);
- for ELOVL4, (5′-GAGCCGGGTAGTGTCCTAAAC-3′ and 5′-CACACGCTTATCTGCGATGG-3′);
- for ELOVL5, (5′-TAACAGGAGTATGGGAAGGCA-3′ and 5′-ACCAGAGGACACGGATAATCTT-3′);
- for ELOVL6, (5′-AACGAGCAAAGTTTGAACTGAGG-3′ and 5′-TCGAAGAGCACCGAATATACTGA-3′);
- for ELOVL7, (5′-GCCTTCAGTGATCTTACATCGAG-3′ and 5′-AGGACATGAGGAGCCAATCTT-3′).
2.4. Statistical Methods
3. Results
3.1. ELOVL1 and ELOVL7 Expression Was Lower in the GBM Tumor Than in the Peritumoral Area
3.2. Expression of Elongases in GBM Tumors Differed between Genders
3.3. Expression of Elongases in the GBM Tumors Correlated with Each Other
3.4. The Expression of Elongases in the GBM Tumors Correlated with BMI and Cigarette Pack-Years in the Men, and Correlated with Age and Body Weight in the Women
3.5. Expression of ELOVL5 and ELOVL6 Increased under the Influence of Hypoxia, and the Expression of ELOVL1, ELOVL3, ELOVL4, and ELOVL7 Decreased
4. Discussion
4.1. Expression of Elongases in GBM Tumors
4.2. Gender Differences in the Expression of Elongases
4.3. Effects of Hypoxia and Nutrient-Deficient Conditions on the Expression of Elongases
4.4. Clinical Significance of Elongases in GBM
4.5. Limitations of the Study
5. Conclusions
- The ELOVL1 and ELOVL7 expressions were reduced in GBM tumors. Very-long-chain SFA synthesis was also reduced in GBM;
- ELOVL2, ELOVL5, ELOVL6, and ELOVL7 expressions were decreased in GBM tumors in women but not in men. The expression of ELOVL1 was upregulated in GBM tumors in women and downregulated in men. The ELOVL4 expression was upregulated in GBM tumors in women. GBM tumors in men showed a higher expression of ELOVL2, ELOVL5, and ELOVL6;
- The demonstrated gender differences in the expression of the studied elongases may allow for the development of personalized therapy targeting fatty acid biosynthesis;
- Hypoxia increased the expression of ELOVL5 and ELOVL6 but decreased the expression of ELOVL1, ELOVL3, ELOVL4, and ELOVL7 in GBM cells. Hypoxia decreased the synthesis of very-long-chain SFA but increased PUFA synthesis;
- Nutrient deficiency reduced the expression of ELOVL1, ELOVL4, ELOVL5, and ELOVL7. Nutrient deficiency decreased the synthesis of very-long-chain SFA but did not affect PUFA synthesis.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grochans, S.; Cybulska, A.M.; Simińska, D.; Korbecki, J.; Kojder, K.; Chlubek, D.; Baranowska-Bosiacka, I. Epidemiology of Glioblastoma Multiforme-Literature Review. Cancers 2022, 14, 2412. [Google Scholar] [CrossRef] [PubMed]
- Mirimanoff, R.O.; Gorlia, T.; Mason, W.; Van den Bent, M.J.; Kortmann, R.D.; Fisher, B.; Reni, M.; Brandes, A.A.; Curschmann, J.; Villa, S.; et al. Radiotherapy and temozolomide for newly diagnosed glioblastoma: Recursive partitioning analysis of the EORTC 26981/22981-NCIC CE3 phase III randomized trial. J. Clin. Oncol. 2006, 24, 2563–2569. [Google Scholar] [CrossRef] [Green Version]
- Lu, V.M.; Goyal, A.; Graffeo, C.S.; Perry, A.; Burns, T.C.; Parney, I.F.; Quinones-Hinojosa, A.; Chaichana, K.L. Survival Benefit of Maximal Resection for Glioblastoma Reoperation in the Temozolomide Era: A Meta-Analysis. World Neurosurg. 2019, 127, 31–37. [Google Scholar] [CrossRef]
- Eriksson, M.; Kahari, J.; Vestman, A.; Hallmans, M.; Johansson, M.; Bergenheim, A.T.; Sandström, M. Improved treatment of glioblastoma—Changes in survival over two decades at a single regional Centre. Acta Oncol. 2019, 58, 334–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantrell, J.N.; Waddle, M.R.; Rotman, M.; Peterson, J.L.; Ruiz-Garcia, H.; Heckman, M.G.; Quiñones-Hinojosa, A.; Rosenfeld, S.S.; Brown, P.D.; Trifiletti, D.M. Progress Toward Long-Term Survivors of Glioblastoma. Mayo Clin. Proc. 2019, 94, 1278–1286. [Google Scholar] [CrossRef]
- Guillou, H.; Zadravec, D.; Martin, P.G.; Jacobsson, A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 2010, 49, 186–199. [Google Scholar] [CrossRef]
- Libertini, L.J.; Smith, S. Synthesis of long chain acyl-enzyme thioesters by modified fatty acid synthetases and their hydrolysis by a mammary gland thioesterase. Arch. Biochem. Biophys. 1979, 192, 47–60. [Google Scholar] [CrossRef]
- Jakobsson, A.; Westerberg, R.; Jacobsson, A. Fatty acid elongases in mammals: Their regulation and roles in metabolism. Prog. Lipid Res. 2006, 45, 237–249. [Google Scholar] [CrossRef]
- Moon, Y.A.; Horton, J.D. Identification of two mammalian reductases involved in the two-carbon fatty acyl elongation cascade. J. Biol. Chem. 2003, 278, 7335–7343. [Google Scholar] [CrossRef]
- Matsuzaka, T.; Shimano, H.; Yahagi, N.; Yoshikawa, T.; Amemiya-Kudo, M.; Hasty, A.H.; Okazaki, H.; Tamura, Y.; Iizuka, Y.; Ohashi, K.; et al. Cloning and characterization of a mammalian fatty acyl-CoA elongase as a lipogenic enzyme regulated by SREBPs. J. Lipid Res. 2002, 43, 911–920. [Google Scholar] [CrossRef]
- Matsuzaka, T.; Shimano, H.; Yahagi, N.; Kato, T.; Atsumi, A.; Yamamoto, T.; Inoue, N.; Ishikawa, M.; Okada, S.; Ishigaki, N.; et al. Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat. Med. 2007, 13, 1193–1202. [Google Scholar] [CrossRef]
- Ohno, Y.; Suto, S.; Yamanaka, M.; Mizutani, Y.; Mitsutake, S.; Igarashi, Y.; Sassa, T.; Kihara, A. ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 18439–18444. [Google Scholar] [CrossRef] [Green Version]
- Naganuma, T.; Sato, Y.; Sassa, T.; Ohno, Y.; Kihara, A. Biochemical characterization of the very long-chain fatty acid elongase ELOVL7. FEBS Lett. 2011, 585, 3337–3341. [Google Scholar] [CrossRef] [Green Version]
- Leonard, A.E.; Bobik, E.G.; Dorado, J.; Kroeger, P.E.; Chuang, L.T.; Thurmond, J.M.; Parker-Barnes, J.M.; Das, T.; Huang, Y.S.; Mukerji, P. Cloning of a human cDNA encoding a novel enzyme involved in the elongation of long-chain polyunsaturated fatty acids. Biochem. J. 2000, 350, 765–770. [Google Scholar] [CrossRef]
- Leonard, A.E.; Kelder, B.; Bobik, E.G.; Chuang, L.T.; Lewis, C.J.; Kopchick, J.J.; Mukerji, P.; Huang, Y.S. Identification and expression of mammalian long-chain PUFA elongation enzymes. Lipids 2002, 37, 733–740. [Google Scholar] [CrossRef]
- Agbaga, M.P.; Brush, R.S.; Mandal, M.N.; Henry, K.; Elliott, M.H.; Anderson, R.E. Role of Stargardt-3 macular dystrophy protein (ELOVL4) in the biosynthesis of very long chain fatty acids. Proc. Natl. Acad. Sci. USA 2008, 105, 12843–12848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hopiavuori, B.R.; Anderson, R.E.; Agbaga, M.P. ELOVL4: Very long-chain fatty acids serve an eclectic role in mammalian health and function. Prog. Retin. Eye Res. 2019, 69, 137–158. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Kojder, K.; Jeżewski, D.; Simińska, D.; Tarnowski, M.; Kopytko, P.; Safranow, K.; Gutowska, I.; Goschorska, M.; Kolasa-Wołosiuk, A.; et al. Expression of SCD and FADS2 Is Lower in the Necrotic Core and Growing Tumor Area than in the Peritumoral Area of Glioblastoma Multiforme. Biomolecules 2020, 10, 727. [Google Scholar] [CrossRef]
- Shabihkhani, M.; Telesca, D.; Movassaghi, M.; Naeini, Y.B.; Naeini, K.M.; Hojat, S.A.; Gupta, D.; Lucey, G.M.; Ontiveros, M.; Wang, M.W.; et al. Incidence, survival, pathology, and genetics of adult Latino Americans with glioblastoma. J. Neurooncol. 2017, 132, 351–358. [Google Scholar] [CrossRef]
- Tian, M.; Ma, W.; Chen, Y.; Yu, Y.; Zhu, D.; Shi, J.; Zhang, Y. Impact of gender on the survival of patients with glioblastoma. Biosci. Rep. 2018, 38, BSR20180752. [Google Scholar] [CrossRef] [Green Version]
- Baid, U.; Talbar, S.; Rane, S.; Gupta, S.; Thakur, M.H.; Moiyadi, A.; Sable, N.; Akolkar, M.; Mahajan, A. A Novel Approach for Fully Automatic Intra-Tumor Segmentation With 3D U-Net Architecture for Gliomas. Front. Comput. Neurosci. 2020, 14, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, X.; Tustison, N.J.; Patel, S.H.; Meyer, C.H. Brain Tumor Segmentation Using an Ensemble of 3D U-Nets and Overall Survival Prediction Using Radiomic Features. Front. Comput. Neurosci. 2020, 14, 25. [Google Scholar] [CrossRef] [Green Version]
- Lemée, J.M.; Com, E.; Clavreul, A.; Avril, T.; Quillien, V.; de Tayrac, M.; Pineau, C.; Menei, P. Proteomic analysis of glioblastomas: What is the best brain control sample? J. Proteomics 2013, 85, 165–173. [Google Scholar] [CrossRef] [Green Version]
- Brat, D.J.; Castellano-Sanchez, A.A.; Hunter, S.B.; Pecot, M.; Cohen, C.; Hammond, E.H.; Devi, S.N.; Kaur, B.; Van Meir, E.G. Pseudopalisades in glioblastoma are hypoxic, express extracellular matrix proteases, and are formed by an actively migrating cell population. Cancer Res. 2004, 64, 920–927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz-Sánchez, J.; Chánez-Cárdenas, M.E. The use of cobalt chloride as a chemical hypoxia model. J. Appl. Toxicol. 2019, 39, 556–570. [Google Scholar] [CrossRef] [PubMed]
- Said, H.M.; Hagemann, C.; Stojic, J.; Schoemig, B.; Vince, G.H.; Flentje, M.; Roosen, K.; Vordermark, D. GAPDH is not regulated in human glioblastoma under hypoxic conditions. BMC Mol. Biol. 2007, 8, 55. [Google Scholar] [CrossRef] [Green Version]
- Seifert, M.; Garbe, M.; Friedrich, B.; Mittelbronn, M.; Klink, B. Comparative transcriptomics reveals similarities and differences between astrocytoma grades. BMC Cancer 2015, 15, 952. [Google Scholar] [CrossRef] [PubMed]
- Saurty-Seerunghen, M.S.; Bellenger, L.; El-Habr, E.A.; Delaunay, V.; Garnier, D.; Chneiweiss, H.; Antoniewski, C.; Morvan-Dubois, G.; Junier, M.P. Capture at the single cell level of metabolic modules distinguishing aggressive and indolent glioblastoma cells. Acta Neuropathol. Commun. 2019, 7, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vyazunova, I.; Maklakova, V.I.; Berman, S.; De, I.; Steffen, M.D.; Hong, W.; Lincoln, H.; Morrissy, A.S.; Taylor, M.D.; Akagi, K.; et al. Sleeping Beauty mouse models identify candidate genes involved in gliomagenesis. PLoS ONE 2014, 9, e113489. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, M.; Koparan, M.; Sari, A.; Ozturk, S.; Kaplan, S.K.; Erol, F.S. Can Behenic Acid (C22:0) Levels be a Prognostic Factor in Glial Tumors? Can J. Neurol. Sci. 2013, 40, 854–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Igarashi, M.; Ma, K.; Chang, L.; Bell, J.M.; Rapoport, S.I. Dietary n-3 PUFA deprivation for 15 weeks upregulates elongase and desaturase expression in rat liver but not brain. J. Lipid Res. 2007, 48, 2463–2470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alessandri, J.M.; Extier, A.; Al-Gubory, K.H.; Langelier, B.; Baudry, C.; LePoupon, C.; Lavialle, M.; Guesnet, P. Ovariectomy and 17β-estradiol alter transcription of lipid metabolism genes and proportions of neo-formed n-3 and n-6 long-chain polyunsaturated fatty acids differently in brain and liver. J. Nutr. Biochem. 2011, 22, 820–827. [Google Scholar] [CrossRef]
- Marks, K.A.; Kitson, A.P.; Stark, K.D. Hepatic and plasma sex differences in saturated and monounsaturated fatty acids are associated with differences in expression of elongase 6, but not stearoyl-CoA desaturase in Sprague-Dawley rats. Genes Nutr. 2013, 8, 317–327. [Google Scholar] [CrossRef] [Green Version]
- Marks, K.A.; Kitson, A.P.; Shaw, B.; Mutch, D.M.; Stark, K.D. Stearoyl-CoA desaturase 1, elongase 6 and their fatty acid products and precursors are altered in ovariectomized rats with 17β-estradiol and progesterone treatment. Prostaglandins Leukot. Essent. Fat. Acids 2013, 89, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Marra, C.A.; de Alaniz, M.J. Influence of testosterone administration on the biosynthesis of unsaturated fatty acids in male and female rats. Lipids 1989, 24, 1014–1019. [Google Scholar] [CrossRef] [PubMed]
- Childs, C.E.; Hoile, S.P.; Burdge, G.C.; Calder, P.C. Changes in rat n-3 and n-6 fatty acid composition during pregnancy are associated with progesterone concentrations and hepatic FADS2 expression. Prostaglandins Leukot. Essent. Fat. Acids 2012, 86, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Kitson, A.P.; Marks, K.A.; Shaw, B.; Mutch, D.M.; Stark, K.D. Treatment of ovariectomized rats with 17β-estradiol increases hepatic delta-6 desaturase enzyme expression and docosahexaenoic acid levels in hepatic and plasma phospholipids. Prostaglandins Leukot. Essent. Fat. Acids 2013, 89, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Sibbons, C.M.; Brenna, J.T.; Lawrence, P.; Hoile, S.P.; Clarke-Harris, R.; Lillycrop, K.A.; Burdge, G.C. Effect of sex hormones on n-3 polyunsaturated fatty acid biosynthesis in HepG2 cells and in human primary hepatocytes. Prostaglandins Leukot. Essent. Fat. Acids 2014, 90, 47–54. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Jandacek, R.; Rider, T.; Tso, P.; McNamara, R.K. Elevated delta-6 desaturase (FADS2) expression in the postmortem prefrontal cortex of schizophrenic patients: Relationship with fatty acid composition. Schizophr. Res. 2009, 109, 113–120. [Google Scholar] [CrossRef]
- Kitson, A.P.; Smith, T.L.; Marks, K.A.; Stark, K.D. Tissue-specific sex differences in docosahexaenoic acid and Δ6-desaturase in rats fed a standard chow diet. Appl. Physiol. Nutr. Metab. 2012, 37, 1200–1211. [Google Scholar] [CrossRef] [PubMed]
- González-Bengtsson, A.; Asadi, A.; Gao, H.; Dahlman-Wright, K.; Jacobsson, A. Estrogen Enhances the Expression of the Polyunsaturated Fatty Acid Elongase Elovl2 via ERα in Breast Cancer Cells. PLoS ONE 2016, 11, e0164241. [Google Scholar] [CrossRef] [Green Version]
- Burger, H. The menopausal transition—Endocrinology. J. Sex. Med. 2008, 5, 2266–2273. [Google Scholar] [CrossRef] [PubMed]
- Idbaih, A.; Aimard, J.; Boisselier, B.; Marie, Y.; Paris, S.; Crinière, E.; Carvalho Silva, R.; Laigle-Donadey, F.; Rousseau, A.; Mokhtari, K.; et al. Epidermal growth factor receptor extracellular domain mutations in primary glioblastoma. Neuropathol. Appl. Neurobiol. 2009, 35, 208–213. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Coleman, W.; Huang, W.; Rubin, J.B.; Lathia, J.D.; Berens, M.E.; Speyer, G.; Liao, P.; Wrensch, M.R.; Eckel-Passow, J.E.; et al. Sex-specific gene and pathway modeling of inherited glioma risk. Neuro-oncology 2019, 21, 71–82. [Google Scholar] [CrossRef] [Green Version]
- An, Z.; Aksoy, O.; Zheng, T.; Fan, Q.W.; Weiss, W.A. Epidermal growth factor receptor and EGFRvIII in glioblastoma: Signaling pathways and targeted therapies. Oncogene 2018, 37, 1561–1575. [Google Scholar] [CrossRef]
- Guo, D.; Prins, R.M.; Dang, J.; Kuga, D.; Iwanami, A.; Soto, H.; Lin, K.Y.; Huang, T.T.; Akhavan, D.; Hock, M.B.; et al. EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Sci. Signal. 2009, 2, ra82. [Google Scholar] [CrossRef] [Green Version]
- Qin, Y.; Dalen, K.T.; Gustafsson, J.A.; Nebb, H.I. Regulation of hepatic fatty acid elongase 5 by LXRalpha-SREBP-1c. Biochim. Biophys. Acta 2009, 1791, 140–147. [Google Scholar] [CrossRef]
- Shikama, A.; Shinozaki, H.; Takeuchi, Y.; Matsuzaka, T.; Aita, Y.; Murayama, T.; Sawada, Y.; Piao, X.; Toya, N.; Oya, Y.; et al. Identification of human ELOVL5 enhancer regions controlled by SREBP. Biochem. Biophys. Res. Commun. 2015, 465, 857–863. [Google Scholar] [CrossRef]
- Moon, Y.A.; Shah, N.A.; Mohapatra, S.; Warrington, J.A.; Horton, J.D. Identification of a mammalian long chain fatty acyl elongase regulated by sterol regulatory element-binding proteins. J. Biol. Chem. 2001, 276, 45358–45366. [Google Scholar] [CrossRef]
- Wang, Y.; Botolin, D.; Xu, J.; Christian, B.; Mitchell, E.; Jayaprakasam, B.; Nair, M.G.; Peters, J.M.; Busik, J.V.; Olson, L.K.; et al. Regulation of hepatic fatty acid elongase and desaturase expression in diabetes and obesity. J. Lipid Res. 2006, 47, 2028–2041. [Google Scholar] [CrossRef] [Green Version]
- Purdy, J.G.; Shenk, T.; Rabinowitz, J.D. Fatty acid elongase 7 catalyzes lipidome remodeling essential for human cytomegalovirus replication. Cell Rep. 2015, 10, 1375–1385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, C.A.; Brault, C.; Peck, B.; Bensaad, K.; Griffiths, B.; Mitter, R.; Chakravarty, P.; East, P.; Dankworth, B.; Alibhai, D.; et al. SREBP maintains lipid biosynthesis and viability of cancer cells under lipid- and oxygen-deprived conditions and defines a gene signature associated with poor survival in glioblastoma multiforme. Oncogene 2015, 34, 5128–5140. [Google Scholar] [CrossRef] [PubMed]
- Furuta, E.; Pai, S.K.; Zhan, R.; Bandyopadhyay, S.; Watabe, M.; Mo, Y.Y.; Hirota, S.; Hosobe, S.; Tsukada, T.; Miura, K.; et al. Fatty acid synthase gene is up-regulated by hypoxia via activation of Akt and sterol regulatory element binding protein-1. Cancer Res. 2008, 68, 1003–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimano, H. Sterol regulatory element-binding proteins (SREBPs): Transcriptional regulators of lipid synthetic genes. Prog. Lipid Res. 2001, 40, 439–452. [Google Scholar] [CrossRef]
- Bougarne, N.; Weyers, B.; Desmet, S.J.; Deckers, J.; Ray, D.W.; Staels, B.; De Bosscher, K. Molecular Actions of PPARα in Lipid Metabolism and Inflammation. Endocr. Rev. 2018, 39, 760–802. [Google Scholar] [CrossRef] [Green Version]
- Kady, N.M.; Liu, X.; Lydic, T.A.; Syed, M.H.; Navitskaya, S.; Wang, Q.; Hammer, S.S.; O’Reilly, S.; Huang, C.; Seregin, S.S.; et al. ELOVL4-Mediated Production of Very Long-Chain Ceramides Stabilizes Tight Junctions and Prevents Diabetes-Induced Retinal Vascular Permeability. Diabetes 2018, 67, 769–781. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; He, Q.; Guo, Z.; Yang, L.; Bao, L.; Bao, W.; Zheng, X.; Wang, Y.; Wang, Z. Inhibition of Mammalian Target of Rapamycin Complex 1 (mTORC1) Downregulates ELOVL1 Gene Expression and Fatty Acid Synthesis in Goat Fetal Fibroblasts. Int. J. Mol. Sci. 2015, 16, 16440–16453. [Google Scholar] [CrossRef] [Green Version]
- Leprivier, G.; Rotblat, B. How does mTOR sense glucose starvation? AMPK is the usual suspect. Cell Death Discov. 2020, 6, 27. [Google Scholar] [CrossRef] [Green Version]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Patil, N.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013-2017. Neuro-oncology 2020, 22, iv96. [Google Scholar] [CrossRef] [PubMed]
- Expression of ELOVL3 in Glioma-The Human Protein Atlas. Version 21.1. Available online: https://www.proteinatlas.org/ENSG00000119915-ELOVL3/pathology/glioma (accessed on 3 September 2022).
- Expression of ELOVL1 in Glioma-The Human Protein Atlas. Version 21.1. Available online: https://www.proteinatlas.org/ENSG00000066322-ELOVL1/pathology/glioma (accessed on 3 September 2022).
- Expression of ELOVL7 in Glioma-The Human Protein Atlas. Version 21.1. Available online: https://www.proteinatlas.org/ENSG00000164181-ELOVL7/pathology/glioma (accessed on 3 September 2022).
- Zhang, Y.; Pang, S.; Sun, B.; Zhang, M.; Jiao, X.; Lai, L.; Qian, Y.; Yang, N.; Yang, W. ELOVLs Predict Distinct Prognosis Value and Immunotherapy Efficacy in Patients With Hepatocellular Carcinoma. Front. Oncol. 2022, 12, 884066. [Google Scholar] [CrossRef]
- Qiu, J.; Shi, Z.; Jiang, J. Cyclooxygenase-2 in glioblastoma multiforme. Drug Discov. Today 2017, 22, 148–156. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Liang, H.; Sun, M.; Zhang, L.; Xu, H.; Liu, W.; Li, Y.; Zhou, Y.; Li, Y.; Li, M. Delta-6-desaturase inhibitor enhances radiation therapy in glioblastoma in vitro and in vivo. Cancer Manag. Res. 2018, 10, 6779–6790. [Google Scholar] [CrossRef] [Green Version]
- Tsai, Y.T.; Lo, W.L.; Chen, P.Y.; Ko, C.Y.; Chuang, J.Y.; Kao, T.J.; Yang, W.B.; Chang, K.Y.; Hung, C.Y.; Kikkawa, U.; et al. Reprogramming of arachidonate metabolism confers temozolomide resistance to glioblastoma through enhancing mitochondrial activity in fatty acid oxidation. J. Biomed. Sci. 2022, 29, 21. [Google Scholar] [CrossRef]
- Expression of ELOVL5 in Glioma-The Human Protein Atlas. Version 21.1. Available online: https://www.proteinatlas.org/ENSG00000012660-ELOVL5/pathology/glioma (accessed on 3 September 2022).
- Gimple, R.C.; Kidwell, R.L.; Kim, L.J.Y.; Sun, T.; Gromovsky, A.D.; Wu, Q.; Wolf, M.; Lv, D.; Bhargava, S.; Jiang, L.; et al. Glioma Stem Cell-Specific Superenhancer Promotes Polyunsaturated Fatty-Acid Synthesis to Support EGFR Signaling. Cancer Discov. 2019, 9, 1248–1267. [Google Scholar] [CrossRef]
- Expression of ELOVL2 in Glioma-The Human Protein Atlas. Version 21.1. Available online: https://www.proteinatlas.org/ENSG00000197977-ELOVL2/pathology/glioma (accessed on 3 September 2022).
- Expression of ELOVL6 in Glioma-The Human Protein Atlas. Version 21.1. Available online: https://www.proteinatlas.org/ENSG00000170522-ELOVL6/pathology/glioma (accessed on 3 September 2022).
- Su, Y.C.; Feng, Y.H.; Wu, H.T.; Huang, Y.S.; Tung, C.L.; Wu, P.; Chang, C.J.; Shiau, A.L.; Wu, C.L. Elovl6 is a negative clinical predictor for liver cancer and knockdown of Elovl6 reduces murine liver cancer progression. Sci. Rep. 2018, 8, 6586. [Google Scholar]
- Expression of ELOVL4 in Glioma-The Human Protein Atlas. Version 21.1. Available online: https://www.proteinatlas.org/ENSG00000118402-ELOVL4/pathology/glioma (accessed on 3 September 2022).





| N | Mean | Standard Deviation | Minimum | Maximum | |
|---|---|---|---|---|---|
| Age at surgery | 24 | 60.7 | 12.5 | 36 | 81 |
| Weight | 24 | 84 | 19 | 55 | 130 |
| Height | 23 | 172 | 12 | 147 | 196 |
| BMI | 23 | 28.7 | 4.8 | 21.5 | 38.9 |
| Pack-years (number of smoking years × number of packs (20 pcs.)/d | 20 | 13.42 | 13.98 | 0 | 42 |
| Men | |||||
| Age at surgery | 14 | 60.6 | 11.9 | 41 | 81 |
| Weight | 14 | 93.8 | 15.4 | 73 | 130 |
| Height | 14 | 178 | 8.6 | 163 | 196 |
| BMI | 14 | 29.5 | 4.2 | 24.7 | 38.9 |
| Pack-years (number of smoking years × number of packs (20 pcs.)/d | 12 | 18.63 | 14.48 | 0 | 42 |
| Women | |||||
| Age at surgery | 12 | 60.8 | 14.1 | 36 | 79 |
| Weight | 12 | 70.3 | 15 | 55 | 95 |
| Height | 11 | 162 | 8 | 147 | 173 |
| BMI | 11 | 27.4 | 5.7 | 21.5 | 36.2 |
| Pack-years (number of smoking years × number of packs (20 pcs.)/d | 8 | 5.61 | 9.23 | 0 | 21 |
| Hormone replacement therapy | 7 | 5 patients were not receiving hormone therapy, 2 patients were receiving | |||
| Studied Gene/ Location | ELOVL1 | ELOVL2 | ELOVL3 | ELOVL4 | ELOVL5 | ELOVL6 | ELOVL7 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETR | TC | PA | ETR | TC | PA | ETR | TC | PA | ETR | TC | PA | ETR | TC | PA | ETR | TC | PA | ETR | TC | PA | ||
| ELOVL1 | ETR | 1.00 | ||||||||||||||||||||
| TC | 0.36 | 1.00 | ||||||||||||||||||||
| PA | −0.32 | 0.07 | 1.00 | |||||||||||||||||||
| ELOVL2 | ETR | 0.35 | −0.27 | 0.34 | 1.00 | |||||||||||||||||
| TC | 0.44 * | 0.17 | 0.04 | 0.02 | 1.00 | |||||||||||||||||
| PA | 0.35 | −0.06 | 0.25 | 0.34 | 0.07 | 1.00 | ||||||||||||||||
| ELOVL3 | ETR | −0.02 | −0.15 | −0.02 | 0.37 | 0.29 | 0.13 | 1.00 | ||||||||||||||
| TC | −0.44 | −0.18 | −0.05 | −0.49 * | 0.16 | −0.33 | −0.03 | 1.00 | ||||||||||||||
| PA | −0.34 | −0.23 | −0.11 | 0.19 | 0.31 | 0.02 | 0.08 | 0.10 | 1.00 | |||||||||||||
| ELOVL4 | ETR | 0.05 | −0.02 | 0.06 | 0.26 | 0.15 | −0.10 | 0.07 | −0.10 | −0.33 | 1.00 | |||||||||||
| TC | −0.09 | −0.14 | 0.04 | −0.04 | 0.37 * | 0.16 | 0.23 | −0.02 | −0.12 | −0.14 | 1.00 | |||||||||||
| PA | −0.06 | 0.35 | −0.07 | −0.09 | 0.46 * | 0.49 * | 0.02 | 0.36 | 0.03 | 0.36 | 0.51 * | 1.00 | ||||||||||
| ELOVL5 | ETR | 0.30 | 0.04 | 0.19 | 0.50 * | −0.14 | 0.25 | 0.00 | 0.53 * | 0.01 | 0.46 * | −0.09 | 0.18 | 1.00 | ||||||||
| TC | 0.26 | 0.58 * | −0.03 | −0.12 | 0.65 * | −0.22 | −0.07 | 0.22 | 0.16 | 0.03 | 0.77 * | 0.35 | 0.08 | 1.00 | ||||||||
| PA | 0.13 | 0.16 | 0.24 | 0.16 | 0.01 | 0.74 * | −0.05 | −0.03 | 0.02 | −0.15 | 0.24 | 0.51 * | 0.10 | 0.17 | 1.00 | |||||||
| ELOVL6 | ETR | 0.36 | 0.05 | 0.03 | 0.34 | −0.22 | 0.25 | 0.22 | 0.32 | −0.01 | 0.53 * | 0.15 | 0.22 | 0.72 * | 0.73 * | −0.13 | 1.00 | |||||
| TC | 0.10 | 0.28 | −0.12 | −0.13 | 0.66 * | −0.04 | 0.08 | 0.23 | 0.13 | 0.03 | 0.64 * | 0.28 | 0.06 | 0.83 * | 0.09 | 0.34 | 1.00 | |||||
| PA | −0.04 | 0.03 | 0.21 | −0.01 | 0.20 | 0.61 * | −0.19 | −0.17 | 0.06 | 0.33 | 0.20 | 0.73 * | 0.07 | 0.26 | 0.51 * | 0.07 | 0.28 | 1.00 | ||||
| ELOVL7 | ETR | −0.12 | 0.17 | 0.00 | 0.22 | −0.03 | −0.02 | 0.26 | 0.25 | −0.26 | 0.60 * | 0.05 | 0.40 * | 0.54 * | −0.03 | −0.08 | 0.51 * | −0.32 | 0.31 | 1.00 | ||
| TC | −0.04 | −0.04 | −0.13 | 0.01 | 0.31 | −0.09 | −0.12 | −0.11 | 0.03 | 0.31 | 0.60 * | −0.23 | 0.29 | 0.70 * | −0.06 | 0.34 | 0.49 * | 0.06 | 0.29 | 1.00 | ||
| PA | −0.08 | −0.13 | 0.24 | −0.01 | 0.33 | 0.58 * | 0.03 | −0.30 | −0.15 | −0.52 * | 0.08 | 0.11 | 0.02 | 0.11 | 0.46 * | −0.01 | 0.21 | 0.58 * | −0.46 * | −0.13 | 1.00 | |
| Studied Gene/ Location | ELOVL1 | ELOVL2 | ELOVL3 | ELOVL4 | ELOVL5 | ELOVL6 | ELOVL7 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETR | TC | PA | ETR | TC | PA | ETR | TC | PA | ETR | TC | PA | ETR | TC | PA | ETR | TC | PA | ETR | TC | PA | ||
| ELOVL1 | ETR | 1.00 | ||||||||||||||||||||
| TC | 0.18 | 1.00 | ||||||||||||||||||||
| PA | 0.15 | 0.06 | 1.00 | |||||||||||||||||||
| ELOVL2 | ETR | 0.32 | −0.51 | 0.13 | 1.00 | |||||||||||||||||
| TC | 0.49 | 0.29 | 0.04 | 0.39 * | 1.00 | |||||||||||||||||
| PA | 0.41 | 0.11 | 0.36 | 0.70 * | 0.46 | 1.00 | ||||||||||||||||
| ELOVL3 | ETR | −0.12 | −0.03 | 0.06 | 0.52 * | 0.28 | 0.10 | 1.00 | ||||||||||||||
| TC | −0.39 | −0.16 | 0.05 | −0.59 * | 0.14 | −0.64 * | −0.35 | 1.00 | ||||||||||||||
| PA | 0.18 | 0.31 | 0.05 | 0.43 | 0.37 | 0.29 | 0.60 * | −0.21 | 1.00 | |||||||||||||
| ELOVL4 | ETR | −0.04 | 0.02 | −0.03 | −0.24 | 0.08 | −0.03 | 0.15 | −0.09 | 0.16 | 1.00 | |||||||||||
| TC | 0.00 | 0.54 * | −0.26 | 0.09 | 0.27 | 0.14 | 0.29 | −0.04 | 0.19 | −0.08 | 1.00 | |||||||||||
| PA | −0.08 | 0.44 | 0.21 | −0.06 | 0.48 * | 0.61 * | 0.24 | −0.37 | 0.32 | 0.55 * | 0.40 | 1.00 | ||||||||||
| ELOVL5 | ETR | 0.39 | −0.24 | 0.25 | 0.40 | 0.36 | 0.59 * | 0.02 | −0.17 | 0.27 | 0.26 | −0.27 | 0.29 | 1.00 | ||||||||
| TC | 0.31 | 0.67 * | −0.02 | −0.11 | 0.67 * | 0.21 | 0.05 | 0.28 | 0.23 | −0.02 | 0.76 * | 0.22 | 0.19 | 1.00 | ||||||||
| PA | 0.19 | 0.25 | 0.36 | 0.40 | 0.35 | 0.68 * | −0.30 | −0.38 | 0.06 | 0.22 | 0.11 | 0.57 * | 0.47 | 0.20 | 1.00 | |||||||
| ELOVL6 | ETR | 0.32 | 0.04 | −0.31 | 0.29 | 0.42 * | 0.41 * | −0.08 | −0.32 | 0.44 | 0.60 * | 0.14 | 0.46 | 0.66 * | 0.32 | 0.26 | 1.00 | |||||
| TC | 0.15 | 0.49 * | 0.14 | −0.09 | 0.65 * | 0.08 | 0.15 | 0.14 | 0.19 | 0.15 | 0.60 * | 0.27 | 0.03 | 0.82 * | 0.16 | 0.31 | 1.00 | |||||
| PA | 0.26 | 0.19 | 0.35 | 0.06 | 0.54 * | 0.72 * | −0.11 | −0.40 | 0.17 | −0.23 | 0.31 | 0.83 * | 0.20 | 0.26 | 0.67 * | 0.09 | 0.22 | 1.00 | ||||
| ELOVL7 | ETR | −0.50 | 0.15 | −0.24 | −0.49 | −0.33 | −0.18 | 0.09 | −0.34 | 0.17 | 0.69 * | 0.16 | 0.64 * | 0.19 | −0.25 | −0.30 | 0.54 * | −0.19 | −0.09 | 1.00 | ||
| TC | 0.20 | 0.69 * | −0.23 | −0.12 | 0.11 | −0.02 | 0.04 | −0.05 | −0.07 | −0.15 | 0.76 * | 0.25 | −0.28 | 0.65 * | −0.01 | 0.07 | 0.43 | 0.08 | −0.07 | 1.00 | ||
| PA | 0.19 | 0.47 | 0.56 * | 0.36 | 0.36 | 0.66 * | 0.30 | −0.69 * | 0.28 | 0.08 | 0.03 | 0.50 * | 0.45 | 0.09 | 0.53 * | 0.32 | 0.17 | 0.58 * | 0.25 | 0.12 | 1.00 | |
| Studied Gene/ Location | ELOVL1 | ELOVL2 | ELOVL3 | ELOVL4 | ELOVL5 | ELOVL6 | ELOVL7 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETR | TC | PA | ETR | TC | PA | ETR | TC | PA | ETR | TC | PA | ETR | TC | PA | ETR | TC | PA | ETR | TC | PA | ||
| ELOVL1 | ETR | 1.00 | ||||||||||||||||||||
| TC | 0.52 | 1.00 | ||||||||||||||||||||
| PA | 0.36 | 0.22 | 1.00 | |||||||||||||||||||
| ELOVL2 | ETR | 0.41 | −0.07 | 0.53 | 1.00 | |||||||||||||||||
| TC | 0.47 | 0.05 | 0.02 | 0.04 | 1.00 | |||||||||||||||||
| PA | 0.20 | −0.31 | 0.28 | 0.10 | 0.10 | 1.00 | ||||||||||||||||
| ELOVL3 | ETR | −0.15 | −0.23 | 0.28 | 0.17 | 0.39 | 0.22 | 1.00 | ||||||||||||||
| TC | −0.23 | −0.17 | −0.20 | −0.41 | 0.67 * | −0.21 | 0.25 | 1.00 | ||||||||||||||
| PA | −0.23 | −0.28 | −0.35 | −0.05 | 0.29 | −0.55 * | 0.05 | 0.61 * | 1.00 | |||||||||||||
| ELOVL4 | ETR | 0.24 | −0.04 | 0.30 | 0.75 * | 0.06 | 0.02 | 0.44 | −0.22 | −0.05 | 1.00 | |||||||||||
| TC | −0.25 | 0.48 | −0.15 | −0.25 | 0.13 | 0.03 | 0.03 | 0.19 | 0.16 | −0.02 | 1.00 | |||||||||||
| PA | −0.15 | 0.70 * | −0.26 | −0.31 | 0.53 | −0.09 | −0.41 | 0.47 | 0.33 | −0.14 | 0.75 * | 1.00 | ||||||||||
| ELOVL5 | ETR | 0.22 | 0.24 | 0.15 | 0.65 * | −0.22 | 0.17 | 0.13 | −0.74 * | −0.35 | 0.53 * | 0.09 | −0.16 | 1.00 | ||||||||
| TC | 0.25 | 0.55 | −0.18 | −0.26 | 0.63 * | −0.31 | 0.05 | 0.31 | −0.04 | −0.27 | 0.93 * | 0.98 * | 0.10 | 1.00 | ||||||||
| PA | 0.20 | 0.24 | 0.09 | −0.33 | 0.50 * | 0.40 | 0.24 | 0.33 | −0.16 | −0.53 | 0.30 | 0.47 | −0.13 | 0.50 * | 1.00 | |||||||
| ELOVL6 | ETR | 0.20 | 0.07 | 0.28 | 0.51 * | −0.14 | 0.32 | 0.27 | −0.60 * | −0.37 | 0.46 | 0.38 | −0.18 | 0.79 * | 0.16 | −0.27 | 1.00 | |||||
| TC | 0.03 | 0.00 | −0.57 | −0.27 | 0.67 * | 0.37 | 0.24 | 0.57 * | 0.11 | −0.41 | 0.55 | 0.58 | −0.23 | 0.73 * | 0.33 | −0.07 | 1.00 | |||||
| PA | 0.17 | −0.26 | −0.12 | −0.29 | 0.49 * | 0.53 * | −0.29 | 0.24 | −0.30 | 0.00 | 0.10 | 0.15 | −0.26 | 0.15 | 0.07 | −0.06 | −0.10 | 1.00 | ||||
| ELOVL7 | ETR | 0.09 | −0.31 | 0.17 | 0.75 * | 0.37 | 0.49 | 0.21 | −0.20 | −0.09 | 0.64 * | −0.23 | −0.09 | 0.60 * | −0.03 | 0.05 | 0.41 | −0.08 | 0.38 | 1.00 | ||
| TC | 0.05 | −0.03 | 0.39 | 0.50 | 0.27 | 0.26 | −0.09 | −0.10 | −0.05 | 0.07 | −0.24 | −0.05 | 0.54 | 0.17 | 0.43 | 0.36 | −0.13 | 0.08 | 0.66 * | 1.00 | ||
| PA | 0.38 | 0.36 | −0.05 | −0.56 | 0.21 | 0.35 | −0.41 | 0.13 | −0.40 | −0.79 * | 0.07 | 0.40 | −0.38 | 0.43 | 0.58 * | −0.25 | −0.07 | 0.59 * | −0.36 | −0.02 | 1.00 | |
| Parameter | Age | Body Weight | Height | BMI | Pack-Years | Age | Body Weight | Height | BMI | Pack-Years | Age | Body Weight | Height | BMI | Pack-Years | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender | All patients | Men | Women | |||||||||||||
| ELOVL1 | PA | −0.04 | −0.33 | 0.07 | 0.34 | 0.39 | −0.02 | −0.45 | 0.05 | −0.31 | 0.18 | −0.13 | −0.67 * | −0.40 | −0.55 | 0.63 |
| ETR | −0.04 | 0.16 | 0.21 | −0.01 | 0.27 | 0.17 | 0.30 | −0.06 | 0.35 | 0.29 | −0.53 | −0.56 | 0.14 | −0.57 | 0.48 | |
| TC | −0.19 | −0.10 | 0.06 | −0.17 | −0.26 | 0.13 | 0.14 | 0.00 | 0.26 | −0.78 * | −0.63 * | −0.75 * | 0.04 | −0.68 * | 0.81 * | |
| ELOVL2 | PA | −0.01 | −0.23 | −0.47 * | 0.26 | −0.16 | 0.02 | 0.20 | −0.29 | 0.51 * | −0.07 | −0.19 | −0.06 | −0.11 | 0.29 | 0.26 |
| ETR | −0.24 | 0.02 | 0.17 | 0.05 | 0.45 * | 0.08 | −0.24 | −0.40 | 0.20 | 0.77 * | −0.46 * | −0.16 | 0.46 | −0.34 | −0.17 | |
| TC | 0.19 | 0.07 | −0.11 | 0.01 | −0.21 | 0.13 | 0.13 | −0.24 | 0.41 * | 0.00 | 0.17 | −0.48 * | −0.11 | −0.32 | −0.35 | |
| ELOVL3 | PA | 0.02 | −0.16 | −0.25 | −0.15 | −0.28 | −0.06 | −0.22 | −0.27 | 0.02 | 0.35 | 0.14 | 0.19 | 0.11 | −0.05 | −0.54 |
| ETR | −0.01 | −0.36 | −0.31 | −0.34 | −0.29 | 0.20 | −0.54 | −0.46 | −0.15 | 0.26 | 0.07 | −0.08 | 0.16 | −0.36 | 0.00 | |
| TC | 0.21 | −0.44 * | −0.35 | −0.46 * | −0.23 | 0.030 | −0.47 | −0.14 | −0.57 * | 0.01 | 0.44 | −0.38 | −0.41 | −0.22 | −0.34 | |
| ELOVL4 | PA | −0.38 | −0.09 | 0.03 | −0.24 | −0.52 * | −0.52 | 0.27 | 0.38 | 0.20 | −0.42 | −0.01 | −0.25 | −0.14 | −0.29 | 0.00 |
| ETR | −0.38 | 0.24 | 0.63 * | −0.26 | 0.01 | −0.05 | 0.26 | 0.55 | −0.17 | −0.12 | −0.49 | −0.29 | 0.24 | −0.47 | −0.15 | |
| TC | −0.12 | −0.41 | −0.42 | −0.32 | −0.40 | −0.04 | −0.30 | −0.41 | 0.01 | −0.69 * | −0.04 | −0.49 | −0.03 | −0.66 | 0.34 | |
| ELOVL5 | PA | −0.16 | −0.23 | −0.32 | 0.05 | −0.06 | 0.08 | 0.27 | 0.11 | 0.35 | 0.23 | 0.15 | −0.12 | −0.41 | 0.05 | 0.37 |
| ETR | −0.36 | 0.43 * | 0.09 | 0.58 * | 0.27 | 0.14 | 0.23 | −0.13 | 0.39 * | 0.23 | −0.61 * | 0.82 * | 0.01 | −0.45 * | 0.15 | |
| TC | −0.05 | 0.17 | 0.14 | −0.19 | 0.00 | −0.08 | 0.10 | −0.19 | 0.30 | −0.33 | 0.05 | −0.42 * | 0.25 | −0.71 * | 0.06 | |
| ELOVL6 | PA | −0.21 | 0.05 | 0.13 | 0.18 | −0.08 | −0.35 | 0.13 | 0.29 | 0.10 | −0.19 | −0.26 | −0.26 | −0.60 | 0.38 | −0.11 |
| ETR | −0.15 | 0.22 | 0.36 | −0.02 | 0.24 | 0.32 | 0.24 | −0.22 | 0.38 * | −0.33 | −0.34 | −0.17 | −0.07 | −0.52 * | 0.22 | |
| TC | 0.25 | 0.02 | −0.15 | −0.12 | −0.11 | 0.29 | −0.20 | −0.30 | −0.01 | −0.09 | 0.29 | −0.05 | 0.00 | −0.21 | −0.23 | |
| ELOVL7 | PA | 0.12 | −0.04 | −0.35 | 0.22 | −0.28 | 0.10 | 0.55 * | −0.07 | 0.76 * | −0.19 | 0.13 | −0.13 | −0.46 | 0.33 | 0.11 |
| ETR | −0.57 * | 0.27 | 0.46 | 0.18 | −0.28 | −0.50 | 0.50 | 0.36 | 0.25 | −0.71 | −0.55 | 0.01 | 0.36 | −0.02 | −0.73 * | |
| TC | −0.10 | 0.40 | 0.45 | 0.05 | −0.15 | −0.04 | 0.12 | 0.02 | 0.12 | −0.78 * | −0.37 | −0.15 | 0.33 | −0.09 | −0.39 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korbecki, J.; Simińska, D.; Jeżewski, D.; Kojder, K.; Tomasiak, P.; Tarnowski, M.; Chlubek, D.; Baranowska-Bosiacka, I. Glioblastoma Multiforme Tumors in Women Have a Lower Expression of Fatty Acid Elongases ELOVL2, ELOVL5, ELOVL6, and ELOVL7 than in Men. Brain Sci. 2022, 12, 1356. https://doi.org/10.3390/brainsci12101356
Korbecki J, Simińska D, Jeżewski D, Kojder K, Tomasiak P, Tarnowski M, Chlubek D, Baranowska-Bosiacka I. Glioblastoma Multiforme Tumors in Women Have a Lower Expression of Fatty Acid Elongases ELOVL2, ELOVL5, ELOVL6, and ELOVL7 than in Men. Brain Sciences. 2022; 12(10):1356. https://doi.org/10.3390/brainsci12101356
Chicago/Turabian StyleKorbecki, Jan, Donata Simińska, Dariusz Jeżewski, Klaudyna Kojder, Patrycja Tomasiak, Maciej Tarnowski, Dariusz Chlubek, and Irena Baranowska-Bosiacka. 2022. "Glioblastoma Multiforme Tumors in Women Have a Lower Expression of Fatty Acid Elongases ELOVL2, ELOVL5, ELOVL6, and ELOVL7 than in Men" Brain Sciences 12, no. 10: 1356. https://doi.org/10.3390/brainsci12101356