Functional Data Analysis in Sport Science: Example of Swimmers’ Progression Curves Clustering
Abstract
:1. Introduction
1.1. Longitudinal Data in Sport
1.2. Detection of Young Athletes
1.3. Functional Data Analysis (FDA)
1.4. Clustering Functional Data
2. Materials and Methods
2.1. Description of the Real Swimming Data Set
2.2. Testing Several Algorithms on Simulated Data Sets
- (i)
- distance-based:
- distclust: An approximation of the distance between curves is defined, and a k-means heuristic is used on individuals using this distance. This method is well designed in the context of sparsely observed functions with irregular measurements [36].
- (ii)
- model-based:
- fitfclust: One of the first algorithms to use a Gaussian mixture model for univariate functions that we briefly describe. This heuristic holds for all following algorithms described as Gaussian mixture methods. Functions are represented using basis functions, and the associated coefficients are supposed to come from Gaussian distributions. Given a number K of different means and covariances parameters corresponding to the K clusters, an EM algorithm is used to estimate the probability of each observational curves to belong to a cluster. When the iterations stop (various stopping criteria exhist), an individual is affected to its most likely cluster. A preliminary step of FPCA can be added to work on lower dimensional vectors and thus speed up the calculations. This method is well designed in the context of sparsely observed functions [37].
- iterSubspace: A non-parametric model based algorithm. This method uses the Karhunen–Loeve expansion of the curves, and perform a k-means algorithm on the scores of FPCA and the mean process. This method can be useful when the Gaussian assumption does not hold but k-means approach can lead to unstable results [38].
- funclust: A Gaussian mixture model based algorithm. This method uses the Karhunen–Loeve expansion of the curves and allows each cluster’s Gaussian parameters to be of different sizes, according to the quantity of variance expressed by the corresponding FPCA. The algorithm also allows different covariance structures between clusters and thus generalizes some methods such as iterSubspace [33].
- funHDDC: A Gaussian mixture model based algorithm. This method presents lots of common characteristics with funclust, but additionally allows clustering of multivariate functions. The algorithm proposes six ways to model covariates structures, especially for the extra-dimension of the FPCA [35].
- fscm: A non-parametric model based algorithm. Each cluster is modeled by a Markov random field, and functions are clustered by shape regardless to the scale. Observation curves are considered as locally-dependent, and a K-nearest neighbors algorithm is used to define the neighborhood structure. Then, an EM algorithm estimates parameters of the model. This method is well designed when the assumption of independence between curves does not hold [38].
- waveclust: A linear Gaussian mixed effect model algorithm. This approach uses a dimension reduction step using wavelet decomposition (rather than classic FPCA). An EM algorithm is used to compute parameters of the model and probabilities to belong to a cluster. This method is well designed for high-dimension curves, when variations such as peaks appears in data, and thus wavelets perform better than splines [29].
2.3. Clustering the Real Swimming Data Set
2.4. Definition of the Simulated Data Sets
3. Results
3.1. Results on Simulated Data
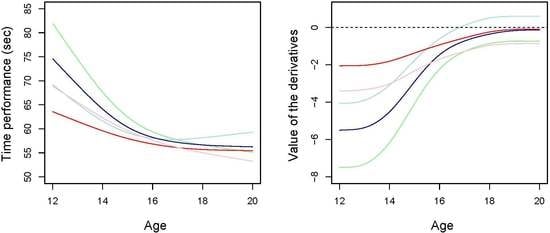
3.2. Data Set of Swimmers’ Progression Curves
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| FDA | Functional Data Analysis |
| FPCA | Functional Principal Component Analysis |
| RI | Rand Index |
| ARI | Adjusted Rand Index |
| BIC | Bayesian Information Criterion |
| ICL | Integrated Classification Likelihood |
| EM | Expectation–Maximization |
Appendix A




References
- Lima-Borges, D.S.; Martinez, P.F.; Vanderlei, L.C.M.; Barbosa, F.S.S.; Oliveira-Junior, S.A. Autonomic Modulations of Heart Rate Variability Are Associated with Sports Injury Incidence in Sprint Swimmers. Phys. Sportsmed. 2018, 46, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Carey, D.L.; Crossley, K.M.; Whiteley, R.; Mosler, A.; Ong, K.L.; Crow, J.; Morris, M.E. Modelling Training Loads and Injuries: The Dangers of Discretization. Med. Sci. Sports Exerc. 2018. [Google Scholar] [CrossRef] [PubMed]
- Boccia, G.; Moisè, P.; Franceschi, A.; Trova, F.; Panero, D.; La Torre, A.; Rainoldi, A.; Schena, F.; Cardinale, M. Career Performance Trajectories in Track and Field Jumping Events from Youth to Senior Success: The Importance of Learning and Development. PLoS ONE 2017, 12, e0170744. [Google Scholar] [CrossRef]
- Brockwell, P.J.; Davis, R.A. Time Series: Theory and Methods; Springer Science & Business Media: Berlin, Germany, 2013. [Google Scholar]
- Warren Liao, T. Clustering of Time Series Data—A Survey. Pattern Recognit. 2005, 38, 1857–1874. [Google Scholar] [CrossRef]
- De Boor, C. On Calculating with B-Splines. J. Approx. Theory 1972, 6, 50–62. [Google Scholar] [CrossRef]
- Forrester, S.E.; Townend, J. The Effect of Running Velocity on Footstrike Angle—A Curve-Clustering Approach. Gait Posture 2015, 41, 26–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mallor, F.; Leon, T.; Gaston, M.; Izquierdo, M. Changes in Power Curve Shapes as an Indicator of Fatigue during Dynamic Contractions. J. Biomech. 2010, 43, 1627–1631. [Google Scholar] [CrossRef] [PubMed]
- Helwig, N.E.; Shorter, K.A.; Ma, P.; Hsiao-Wecksler, E.T. Smoothing Spline Analysis of Variance Models: A New Tool for the Analysis of Cyclic Biomechanical Data. J. Biomech. 2016, 49, 3216–3222. [Google Scholar] [CrossRef] [PubMed]
- Liebl, D.; Willwacher, S.; Hamill, J.; Brüggemann, G.P. Ankle Plantarflexion Strength in Rearfoot and Forefoot Runners: A Novel Clusteranalytic Approach. Hum. Mov. Sci. 2014, 35, 104–120. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, J.O.; Dalzell, C.J. Some Tools for Functional Data Analysis. J. R. Stat. Soc. Ser. B (Methodol.) 1991, 53, 539–572. [Google Scholar]
- Gasser, T.; Muller, H.G.; Kohler, W.; Molinari, L.; Prader, A. Nonparametric Regression Analysis of Growth Curves. Ann. Stat. 1984, 12, 210–229. [Google Scholar] [CrossRef]
- Liebl, D. Modeling and Forecasting Electricity Spot Prices: A Functional Data Perspective. Ann. Appl. Stat. 2013, 7, 1562–1592. [Google Scholar] [CrossRef]
- Bouveyron, C.; Bozzi, L.; Jacques, J.; Jollois, F.X. The Functional Latent Block Model for the Co-Clustering of Electricity Consumption Curves. J. R. Stat. Soc. Ser. C (Appl. Stat.) 2018, 67, 897–915. [Google Scholar] [CrossRef]
- Shen, M.; Tan, H.; Zhou, S.; Smith, G.N.; Walker, M.C.; Wen, S.W. Trajectory of Blood Pressure Change during Pregnancy and the Role of Pre-Gravid Blood Pressure: A Functional Data Analysis Approach. Sci. Rep. 2017, 7, 6227. [Google Scholar] [CrossRef] [PubMed]
- Velasco Herrera, V.M.; Soon, W.; Velasco Herrera, G.; Traversi, R.; Horiuchi, K. Generalization of the Cross-Wavelet Function. New Astron. 2017, 56, 86–93. [Google Scholar] [CrossRef]
- Johnston, K.; Wattie, N.; Schorer, J.; Baker, J. Talent Identification in Sport: A Systematic Review. Sports Med. 2018, 48, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Berthelot, G.; Sedeaud, A.; Marck, A.; Antero-Jacquemin, J.; Schipman, J.; Saulière, G.; Marc, A.; Desgorces, F.D.; Toussaint, J.F. Has Athletic Performance Reached Its Peak? Sports Med. 2015, 45, 1263–1271. [Google Scholar] [CrossRef] [PubMed]
- Moesch, K.; Elbe, A.M.; Hauge, M.L.T.; Wikman, J.M. Late Specialization: The Key to Success in Centimeters, Grams, or Seconds (Cgs) Sports. Scand. J. Med. Sci. Sports 2011, 21, e282–e290. [Google Scholar] [CrossRef] [PubMed]
- Vaeyens, R.; Lenoir, M.; Williams, A.M.; Philippaerts, R.M. Talent Identification and Development Programmes in Sport. Sports Med. 2008, 38, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, H.; Vaeyens, R.; Matthys, S.; Multael, M.; Lefevre, J.; Lenoir, M.; Philippaerts, R. Anthropometric and Performance Measures for the Development of a Talent Detection and Identification Model in Youth Handball. J. Sports Sci. 2009, 27, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Goto, H.; Morris, J.G.; Nevill, M.E. Influence of Biological Maturity on the Match Performance of 8 to 16 Year Old Elite Male Youth Soccer Players. J. Strength Cond. Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Wattie, N.; Schorer, J.; Baker, J. The Relative Age Effect in Sport: A Developmental Systems Model. Sports Med. 2015, 45, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Kearney, P.E.; Hayes, P.R. Excelling at Youth Level in Competitive Track and Field Athletics Is Not a Prerequisite for Later Success. J. Sports Sci. 2018, 36, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Vaeyens, R.; Güllich, A.; Warr, C.R.; Philippaerts, R. Talent Identification and Promotion Programmes of Olympic Athletes. J. Sports Sci. 2009, 27, 1367–1380. [Google Scholar] [CrossRef] [PubMed]
- Ericsson, K.A.; Hoffman, R.R.; Kozbelt, A.; Williams, A.M. The Cambridge Handbook of Expertise and Expert Performance; Cambridge University Press: Cambridge, UK, 2018. [Google Scholar]
- Ramsay, J.O.; Silverman, B.W. Functional Data Analysis; Springer: Berlin, Germany, 2005. [Google Scholar]
- Jacques, J.; Preda, C. Functional Data Clustering: A Survey. Adv. Data Anal. Classif. 2014, 8, 231–255. [Google Scholar] [CrossRef]
- Giacofci, M.; Lambert-Lacroix, S.; Marot, G.; Picard, F. Wavelet-Based Clustering for Mixed-Effects Functional Models in High Dimension. Biometrics 2013, 69, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, J.O.; Silverman, B.W. Applied Functional Data Analysis: Methods and Case Studies; Springer: New York, NY, USA, 2002; Volume 77. [Google Scholar]
- Abraham, C.; Cornillon, P.A.; Matzner-Løber, E.; Molinari, N. Unsupervised Curve Clustering Using B-Splines. Scand. J. Stat. 2003, 30, 581–595. [Google Scholar] [CrossRef]
- Ferraty, F.; Vieu, P. Nonparametric Functional Data Analysis: Theory and Practice; Springer Science & Business Media: Berlin, Germany, 2006. [Google Scholar]
- Jacques, J.; Preda, C. Funclust: A Curves Clustering Method Using Functional Random Variables Density Approximation. Neurocomputing 2013, 112, 164–171. [Google Scholar] [CrossRef]
- Schmutz, A.; Jacques, J.; Bouveyron, C.; Cheze, L.; Martin, P. Clustering Multivariate Functional Data in Group-Specific Functional Subspaces. 2018. Available online: https://hal.inria.fr/hal-01652467/ (accessed on 30 September 2018). HAL.
- Bouveyron, C.; Jacques, J. Model-Based Clustering of Time Series in Group-Specific Functional Subspaces. Adv. Data Anal. Classif. 2011, 5, 281–300. [Google Scholar] [CrossRef]
- Peng, J.; Müller, H.G. Distance-Based Clustering of Sparsely Observed Stochastic Processes, with Applications to Online Auctions. Ann. Appl. Stat. 2008, 2, 1056–1077. [Google Scholar] [CrossRef]
- James, G.M.; Sugar, C.A. Clustering for Sparsely Sampled Functional Data. J. Am. Stat. Assoc. 2003, 98, 397–408. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Serban, N. Clustering Random Curves Under Spatial Interdependence With Application to Service Accessibility. Technometrics 2012, 54, 108–119. [Google Scholar] [CrossRef]
- Rand, W.M. Objective Criteria for the Evaluation of Clustering Methods. J. Am. Stat. Assoc. 1971, 66, 846–850. [Google Scholar] [CrossRef] [Green Version]
- Arbelaitz, O.; Gurrutxaga, I.; Muguerza, J.; Pérez, J.M.; Perona, I.N. An Extensive Comparative Study of Cluster Validity Indices. Pattern Recognit. 2013, 46, 243–256. [Google Scholar] [CrossRef]
- Von Luxburg, U.; Williamson, R.C.; Guyon, I. Clustering: Science or Art? In Proceedings of the ICML Workshop on Unsupervised and Transfer Learning, Bellevue, WA, USA, 2 July 2011; pp. 65–79. [Google Scholar]






| Data Set | Functions | Noise | Observations on t-Axis |
|---|---|---|---|
| Sample 1 | 10 points at regular instants | ||
| Sample 2 | 10 points at regular instants | ||
| Sample 3 | ≤ 10 points at irregular instants |
| Method | Sample 1 | Sample 2 | Sample 3 | Running Speed |
|---|---|---|---|---|
| fitfclust | 0.945 (0.14) | 0.857 (0.01) | 0.307 (0.06) | 2.8 |
| distclust | 0.996 (0.01) | 0.888 (0.05) | 0.523 (0.07) | 19.2 |
| iterSubspace | 0.938 (0.14) | 0.850 (0.12) | 0.527 (0.07) | 1 |
| funclust | 0.450 (0.17) | 0.418 (0.16) | 0.084 (0.07) | 1 |
| fscm | 0.948 (0.12) | 0.902 (0.01) | 0.527 (0.07) | 7 |
| waveclust | 0.920 (0.12) | 0.810 (0.01) | 0.324 (0.13) | 34 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leroy, A.; MARC, A.; DUPAS, O.; REY, J.L.; Gey, S. Functional Data Analysis in Sport Science: Example of Swimmers’ Progression Curves Clustering. Appl. Sci. 2018, 8, 1766. https://doi.org/10.3390/app8101766
Leroy A, MARC A, DUPAS O, REY JL, Gey S. Functional Data Analysis in Sport Science: Example of Swimmers’ Progression Curves Clustering. Applied Sciences. 2018; 8(10):1766. https://doi.org/10.3390/app8101766
Chicago/Turabian StyleLeroy, Arthur, Andy MARC, Olivier DUPAS, Jean Lionel REY, and Servane Gey. 2018. "Functional Data Analysis in Sport Science: Example of Swimmers’ Progression Curves Clustering" Applied Sciences 8, no. 10: 1766. https://doi.org/10.3390/app8101766