Carotenoid Accumulation and the Expression of Carotenoid Metabolic Genes in Mango during Fruit Development and Ripening
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Determination of Fruit Color
2.3. Determination of Ethylene Production and Respiration Rate
2.4. Analysis of Carotenoid Content and Composition
2.5. Isolation and Sequence Analysis of Genes Involved in Carotenoid Metabolism Pathway
2.6. Real Time Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
2.7. Data Analysis
3. Results
3.1. Changes in Respiration Rate, Ethylene Production, Peel and Pulp Color during Mango Fruit Development and Ripening
3.2. Carotenoid Content and Composition in Mango Peel during Fruit Development and Ripening
3.3. Carotenoid Content and Composition in Mango Pulp during Fruit Development and Ripening
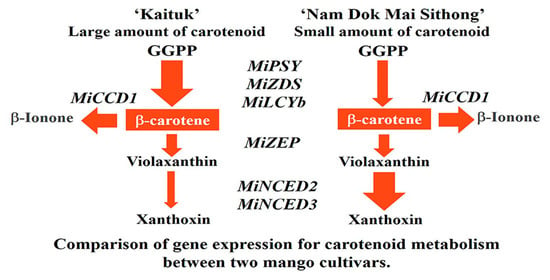
3.4. The Expression of Genes Involved in Carotenoid Metabolism Pathway in Mango Peel during Fruit Development and Ripening
3.5. The Expression of Genes Involved in Carotenoid Metabolism Pathway in Mango Pulp during Fruit Development and Ripening
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ornelas-Paz, D.; Yahia, J.; Gardea, A. Changes in external and internal color during postharvest ripening of ‘Manila’and ‘Ataulfo’mango fruit and relationship with carotenoid content determined by liquid chromatography–APcI+-time-of-flight mass spectrometry. Postharvest Biol. Technol. 2008, 50, 145–152. [Google Scholar] [CrossRef]
- Cooperstone, J.L.; Schwartz, S.J. Recent Insights into Health Benefits of Carotenoids. In Handbook on Natural Pigments in Food and Beverages; Woodhead Publishing: Cambridge, UK, 2016; pp. 473–497. [Google Scholar]
- Tadmor, Y.; Burger, J.; Yaakov, I.; Feder, A.; Libhaber, S.E.; Portnoy, V.; Meir, A.; Tzuri, G.; Sa’ar, U.; Rogachev, I.; et al. Genetics of flavonoid, carotenoid, and chlorophyll pigments in melon fruit rinds. J. Agric. Food Chem. 2010, 58, 10722–10728. [Google Scholar] [CrossRef] [PubMed]
- Kato, M. Mechanism of carotenoid accumulation in citrus fruit. J. Jpn. Soc. Hortic. Sci. 2012, 81, 219–233. [Google Scholar] [CrossRef] [Green Version]
- Liang, M.; Su, X.; Yang, Z.; Deng, H.; Yang, Z.; Liang, R.; Huang, J. Carotenoid composition and expression of carotenogenic genes in the peel and pulp of commercial mango fruit cultivars. Sci. Hortic. 2020, 263, 109072. [Google Scholar] [CrossRef]
- Delgado-Pelayo, R.; Gallardo-Guerrero, L.; Hornero-Méndez, D. Chlorophyll and carotenoid pigments in the peel and flesh of commercial apple fruit varieties. Food Res. Int. 2014, 65, 272–281. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Uribe, L.; Guzman, I.; Rajapakse, W.; Richins, R.D.; O’Connell, M.A. Carotenoid accumulation in orange-pigmented Capsicum annuum fruit, regulated at multiple levels. J. Exp. Bot. 2012, 63, 517–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, H.; Chen, M.; Wen, Q.; Li, Y. Isolation and characterization of the carotenoid biosynthetic genes LCYB, LCYE and CHXB from strawberry and their relation to carotenoid accumulation. Sci. Hortic. 2015, 182, 134–144. [Google Scholar] [CrossRef]
- Lado, J.; Zacarías, L.; Rodrigo, M.J. Regulation of Carotenoid Biosynthesis during Fruit Development. In Carotenoids in Nature; Springer: New York, NY, USA, 2016; pp. 161–198. [Google Scholar]
- Kato, M.; Ikoma, Y.; Matsumoto, H.; Sugiura, M.; Hyodo, H.; Yano, M. Accumulation of Carotenoids and Expression of Carotenoid Biosynthetic Genes during Maturation in Citrus. Plant Physiol. 2004, 134, 824–837. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Yuan, B.; Zhang, M.; Wang, L.; Cui, M.; Wang, Q.; Leng, P. Fruit-specific RNAi-mediated suppression of SlNCED1 increases both lycopene and β-carotene contents in tomato fruit. J. Exp. Bot. 2012, 63, 3097–3108. [Google Scholar] [CrossRef]
- Beekwilder, J.; Van Der Meer, I.M.; Simic, A.; Uitdewilligen, J.; Van Arkel, J.; De Vos, R.C.; Jonker, H.; Verstappen, F.W.; Bouwmeester, H.J.; Sibbesen, O.; et al. Metabolism of carotenoids and apocarotenoids during ripening of raspberry fruit. Biofactors 2008, 34, 57–66. [Google Scholar]
- García-Limones, C.; Schnábele, K.; Blanco-Portales, R.; Luz Bellido, M.; Caballero, J.L.; Schwab, W.; Muñoz-Blanco, J. Functional characterization of FaCCD1: A carotenoid cleavage dioxygenase from strawberry involved in lutein degradation during fruit ripening. J. Agric. Food Chem. 2008, 56, 9277–9285. [Google Scholar] [CrossRef]
- Ampomah-Dwamena, C.; Dejnoprat, S.; Lewis, D.; Sutherland, P.; Volz, R.K.; Allan, A.C. Metabolic and gene expression analysis of apple (Malus × domestica) carotenogenesis. J. Exp. Bot. 2012, 63, 4497–4511. [Google Scholar] [CrossRef]
- Young, P.R.; Lashbrooke, J.G.; Alexandersson, E.; Jacobson, D.; Moser, C.; Velasco, R.; Vivier, M.A. The genes and enzymes of the carotenoid metabolic pathway in Vitis vinifera L. BMC Genom. 2012, 13, 243. [Google Scholar] [CrossRef] [Green Version]
- Ikoma, Y.; Yano, M.; Ogawa, K.; Yoshioka, T.; Xu, Z.C.; Hisada, S.; Omura, M.; Moriguchi, T. Isolation and Evaluation of RNA from Polysaccharide-rich Tissues in Fruit for Quality by cDNA Library Construction and RT-PCR. Jpn. Soc. Hortic. Sci. 1996, 64, 809–814. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Ma, G.; Kato, M.; Yamawaki, K.; Takagi, T.; Kiriiwa, Y.; Ikoma, Y.; Matsumoto, H.; Yoshioka, T.; Nesumi, H. Regulation of Carotenoid Accumulation and the Expression of Carotenoid Metabolic Genes in Citrus Juice Sacs in vitro. J. Exp. Bot. 2012, 63, 871–886. [Google Scholar] [CrossRef] [Green Version]
- Bramley, P.M. Carotenoid Biosynthesis and Chlorophyll Degradation. In The Molecular Biology and Biochemistry of Fruit Ripening; John Wiley & Sons: New York, NY, USA, 2013; pp. 75–116. [Google Scholar]
- Ohmiya, A.; Kato, M.; Shimada, T.; Nashima, K.; Kishimoto, S.; Nagata, M. Molecular basis of carotenoid accumulation in horticultural crops. Hortic. J. 2019, 88, 135–149. [Google Scholar] [CrossRef] [Green Version]
- Barry, C.S.; Giovannoni, J.J. Ethylene and fruit ripening. J. Plant Growth Regul. 2007, 26, 143–159. [Google Scholar] [CrossRef]
- Yin, X.R.; Chen, K.S.; Allan, A.C.; Wu, R.M.; Zhang, B.; Lallu, N.; Ferguson, I.B. Ethylene-induced modulation of genes associated with the ethylene signalling pathway in ripening kiwifruit. J. Exp. Bot. 2008, 59, 2097–2108. [Google Scholar] [CrossRef] [Green Version]
- Honda, C.; Bessho, H.; Murai, M.; Iwanami, H.; Moriya, S.; Abe, K.; Wada, M.; Moriya-Tanaka, Y.; Hayama, H.; Tatsuki, M. Effect of temperature on anthocyanin synthesis and ethylene production in the fruit of early-and medium-maturing apple cultivars during ripening stages. HortScience 2014, 49, 1510–1517. [Google Scholar] [CrossRef]
- Charoenchongsuk, N.; Matsumoto, D.; Itai, A.; Murayama, H. Ripening characteristics and pigment changes in russeted pear fruit in response to ethylene and 1-MCP. Horticulturae 2018, 4, 22. [Google Scholar] [CrossRef] [Green Version]
- Amorós, A.; Zapata, P.; Pretel, M.T.; Botella, M.A.; Serrano, M. Physico-chemical and physiological changes during fruit development and ripening of five loquat (Eriobotrya japonica Lindl.) cultivars. Food Sci. Technol. Int. 2003, 9, 43–51. [Google Scholar] [CrossRef]
- Abbas, M.E.F.; Fandi, B.S. Respiration rate, ethylene production and biochemical changes during fruit development and maturation of jujube (Ziziphus mauritiana Lamk). J. Sci. Food Agric. 2002, 82, 1472–1476. [Google Scholar] [CrossRef]
- Rodriguez-Concepcion, M.; Stange, C. Biosynthesis of carotenoids in carrot: An underground story comes to light. Arch. Biochem. Biophys. 2013, 539, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.H.; Xu, C.J.; Sun, C.D.; Li, X.; Chen, K.S. Carotenoids in white-and red-fleshed loquat fruits. J. Agric. Food Chem. 2007, 55, 7822–7830. [Google Scholar] [CrossRef]
- Feder, A.; Chayut, N.; Gur, A.; Freiman, Z.; Tzuri, G.; Meir, A.; Gal-On, A.; Shnaider, Y.; Wolf, D.; Katzir, N.; et al. The role of the carotenogenic metabolic flux in carotenoid accumulation and chromoplast differentiation: Lessons from the melon fruit. Front. Plant Sci. 2019, 10, 1250. [Google Scholar] [CrossRef]
- Qi, Y.; Liu, X.; Zhang, Q.; Wu, H.; Yan, D.; Liu, Y.; Zhu, X.; Ren, X.; Yang, Y. Carotenoid accumulation and gene expression in fruit skins of three differently colored persimmon cultivars during fruit growth and ripening. Sci. Hortic. 2019, 248, 282–290. [Google Scholar] [CrossRef]
- Yuan, H.; Zhang, J.; Nageswaran, D.; Li, L. Carotenoid Metabolism and Regulation in Horticultural Crops. Hortic. Res. 2015, 2, 15036. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Zheng, B.; Ma, Y.; Xu, W.; Wu, H.; Wang, S. Carotenoid accumulation and expression of carotenoid biosynthesis genes in mango flesh during fruit development and ripening. Sci. Hortic. 2018, 237, 201–206. [Google Scholar] [CrossRef]
- Matile, P.; Hortensteiner, S.; Thomas, H. Chlorophyll degradation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 67–95. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.Y.; Pan, D.L.; Jia, Z.H.; Wang, T.; Wang, G.; Guo, Z.R. Chlorophyll, carotenoid and vitamin C metabolism regulation in Actinidia chinensis’ Hongyang’outer pericarp during fruit development. PLoS ONE 2018, 13, e0194835. [Google Scholar]
- Bramley, P.M. Regulation of carotenoid formation during tomato fruit ripening and development. J. Exp. Bot. 2002, 53, 2107–2113. [Google Scholar] [CrossRef] [Green Version]
- Khoo, H.E.; Ismail, A.; Mohd-Esa, N.; Idris, S. Carotenoid content of underutilized tropical fruits. Plant Food Hum. Nutr. 2008, 63, 170–175. [Google Scholar] [CrossRef]
- Sagawa, J.M.; Stanley, L.E.; LaFountain, A.M.; Frank, H.A.; Liu, C.; Yuan, Y.W. An R2R3MYB transcription factor regulates carotenoid pigmentation in Mimulus lewisii flowers. New Phytol. 2016, 209, 1049–1057. [Google Scholar] [CrossRef]
- Cazzonelli, C.I.; Pogson, B.J. Source to Sink: Regulation of Carotenoid Biosynthesis in Plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef]
- Alquezar, B.; Rodrigo, M.J.; Zacarías, L. Carotenoid biosynthesis and their regulation in citrus fruits. Tree For. Sci. Biotech. 2008, 2, 23–37. [Google Scholar]
- Shen, Y.H.; Yang, F.Y.; Lu, B.G.; Zhao, W.W.; Jiang, T.; Feng, L.; Ming, R. Exploring the differential mechanisms of carotenoid biosynthesis in the yellow peel and red flesh of papaya. BMC Genom. 2019, 20, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Van Eck, J.; Zhou, X. The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation. Plant Cell. 2006, 18, 3594–3605. [Google Scholar] [CrossRef] [Green Version]
- Stanley, L.; Yuan, Y.W. Transcriptional regulation of carotenoid biosynthesis in plants: So many regulators, so little consensus. Front. Plant Sci. 2019, 10, 1017. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Shao, Z.; Zhang, M.; Wang, Q. Regulation of carotenoid metabolism in tomato. Mol. Plant. 2015, 8, 28–39. [Google Scholar] [CrossRef] [Green Version]
- Mlalazi, B.; Welsch, R.; Namanya, P.; Khanna, H.; Geijskes, R.J.; Harrison, M.D.; Harding, R.; Dale, J.L.; Bateson, M. Isolation and functional characterisation of banana phytoenesynthase genes as potential cis-genes. Planta 2012, 236, 1585–1598. [Google Scholar] [CrossRef] [Green Version]
- Dong, T.; Hu, Z.; Deng, L.; Wang, Y.; Zhu, M.; Zhang, J.; Chen, G. A tomato MADS-box transcription factor, SlMADS1, acts as a negative regulator of fruit ripening. Plant Physiol. 2013, 163, 1026–1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marty, I.; Bureau, S.; Sarkissian, G.; Gouble, B.; Audergon, J.M.; Albagnac, G. Ethylene regulation of carotenoid accumulation and carotenogenic gene expression in colour-contrasted apricot varieties (Prunus armeniaca). J. Exp. Bot. 2005, 56, 1877–1886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Ma, G.; Shirai, Y.; Kato, M.; Yamawaki, K.; Ikoma, Y.; Matsumoto, H. Expression and functional analysis of two lycopene β-cyclases from citrus fruits. Planta 2012, 236, 1315–1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, G.; Zhang, L.; Yungyuen, W.; Tsukamoto, I.; Iijima, N.; Oikawa, M.; Kato, M. Expression and functional analysis of citrus carotene hydroxylases: Unravelling the xanthophyll biosynthesis in citrus fruits. BMC Plant Biol. 2016, 16, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.; Kong, W.; Peng, G.; Zhou, J.; Azam, M.; Xu, C.; Grierson, D.; Chen, K. Plastid structure and carotenogenic gene expression in red-and white-fleshed loquat (Eriobotrya japonica) fruits. J. Exp. Bot. 2012, 63, 341–354. [Google Scholar] [CrossRef] [Green Version]
- Ampomah-Dwamena, C.; McGhie, T.; Wibisono, R.; Montefiori, M.; Hellens, R.P.; Allan, A.C. The kiwifruit lycopene beta-cyclase plays a significant role in carotenoid accumulation in fruit. J. Exp. Bot. 2009, 60, 3765–3779. [Google Scholar] [CrossRef] [Green Version]
- Kato, M.; Matsumoto, H.; Ikoma, Y.; Okuda, H.; Yano, M. The role of carotenoid cleavage dioxygenases in the regulation of carotenoid profiles during maturation in citrus fruit. J. Exp. Bot. 2006, 57, 2153–2164. [Google Scholar] [CrossRef] [Green Version]
- Gayen, D.; Ali, N.; Sarkar, S.N.; Datta, S.K.; Datta, K. Down-regulation of lipoxygenase gene reduces degradation of carotenoids of golden rice during storage. Planta 2015, 242, 353–363. [Google Scholar] [CrossRef]
- Ohmiya, A.; Kishimoto, S.; Aida, R.; Yoshioka, S.; Sumitomo, K. Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol. 2006, 142, 1193–1201. [Google Scholar] [CrossRef] [Green Version]
- Brandi, F.; Bar, E.; Mourgues, F.; Horváth, G.; Turcsi, E.; Giuliano, G.; Liverani, A.; Tartarini, S.; Lewinsohn, E.; Rosati, C. Study of ‘Red haven’ peach and its white-fleshed mutant suggests a key role of CCD4 carotenoid dioxygenase in carotenoid and nor isoprenoid volatile metabolism. BMC Plant Biol. 2011, 11, 24. [Google Scholar] [CrossRef] [Green Version]








| cDNA | Forward Primer (F) and Reverse Primer (R) 5’- 3’ | Length (bp) |
|---|---|---|
| MiPSY | F: GTGGTATTGAAGCAGGCAGCCTTGGTTA R: GCATAGGAGACTGGCAATGCAGCTATCT | 943 |
| MiPDS | F: TCTGGCAGATGCAGGCCACAAACCT R: CTGTTGTCACACGCTCAGGTACTCC | 448 |
| MiZDS | F: AGAAACGCTCTGGCTCTTGCTCTAAGTC R: CACTGTGACAACAGGCACGCCAACTAAT | 550 |
| MiCRTISO | F: TGTTAAAGCTGAGGTTCTGCCACCCGAT R: ACAGCTATAACGCCTTGTCCTGGAAACG | 449 |
| MiLCYb | F: CTGGTCTGGCGCAGTTGTTTACATTGAC R: CCGCGCTACAAGTGAAGTTTCCTCAAGA | 483 |
| MiLCYe | F: AGTTGTGAGATCACTGTCAGAGGCTCCA R: CTCCTTCCAGCAATCCGATATAGAACCG | 477 |
| MiHYb | F: GCGTGGCTGAGAAGTTAGCGAGGAA R: CTTCTCTGGGTCTGTGGTGAGACTC | 284 |
| MiZEP | F: CAAGCAAAAGCTCTAGTGAGGAACTGGC R: GCAGAAGTGATCCGACAAGATTTGCACG | 360 |
| MiCCD1 | F: CTGGAGAATCCAGATCTGGACATGGTCA R: GTAATTCCACACTGCAACAGGATCTGGC | 491 |
| MiNCED2 | F: CGAACGTTCCATGAAAACCGTACGGTAC R: CACTCTCCTTCAGTCCTTCATTTCCCGA | 1578 |
| MiNCED3 | F: GCATGGCCAGGTCTTAAACCCAGTTCTA R: CAAGGCAGCCTAAGGATAGCTTTACCTC | 1732 |
| cDNA | TaqMan MGB Probes | Primer Sequences |
|---|---|---|
| MiPSY | ACAGGCAACGACAGAGA | F: CATGGGAATTGCACCTGGAT R: GCCAAGGCAGCATTGTAGACA |
| MiPDS | TGCTATTGGACTTCTTC | F: TTGGCCCGAGAAAGTCAAGT R: GATTGTCCACCAAGCATTGCT |
| MiZDS | TCCCTGGAATTAAAAGAT | F: TGCATATGTTGCTGCATGTGAT R: CCCTCCACGACAATGGAAGT |
| MiCRTISO | CTATGATGCAAAGAAGG | F: GGCAGGGAATGTCTCAAAAGG R: CCAATTATTTCATCTGCCACAAGT |
| MiLCYb | TGATGCAACTGGGTTTT | F: ACAATTCAGGCTGCTGTGGTT R: GCTTATCATACTGAACAAGGCACCTA |
| MiLCYe | TTTCTTCCGGTTACCCAAA | F: GGCATCAGGACATTTTTCCATT R: CGAGAAATCCCTGCCACATC |
| MiHYb | CTCTCTCTGAAATGTTCG | F: CAAATGGAGGGTGGTGAGGTT R: CAGCACCAACAGAGAGTGCAA |
| MiZEP | ATGCCTGGCACCCAA | F: GAGGAAGAGTTGTGCTGGTAGGA R: CCTTGCCCCAGATTTGGA |
| MiCCD1 | TTGGAGGAAATGTTAAAGG | F: GGCTGGAAAGACAAAGCTTGA R: TGGGCCCAGGTCGAAGAT |
| MiNCED2 | ATGAAGTTGTTGTTATAGGATC | F: GGGAGGAGCCGGAATCC R: GTCAGCCGGTGTCATGCA |
| MiNCED3 | TGATAGCTCACCCAAAAG | F: CGACGGTCAGCTTAATTCAACA R: TCGCCTGAAACGGGATCA |
| 18s | Proprietary (Applied Biosystems, Foster City, CA, USA) | Proprietary (Applied Biosystems) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yungyuen, W.; Vo, T.T.; Uthairatanakij, A.; Ma, G.; Zhang, L.; Tatmala, N.; Kaewsuksaeng, S.; Jitareerat, P.; Kato, M. Carotenoid Accumulation and the Expression of Carotenoid Metabolic Genes in Mango during Fruit Development and Ripening. Appl. Sci. 2021, 11, 4249. https://doi.org/10.3390/app11094249
Yungyuen W, Vo TT, Uthairatanakij A, Ma G, Zhang L, Tatmala N, Kaewsuksaeng S, Jitareerat P, Kato M. Carotenoid Accumulation and the Expression of Carotenoid Metabolic Genes in Mango during Fruit Development and Ripening. Applied Sciences. 2021; 11(9):4249. https://doi.org/10.3390/app11094249
Chicago/Turabian StyleYungyuen, Witchulada, Thi Thuong Vo, Apiradee Uthairatanakij, Gang Ma, Lancui Zhang, Nopparat Tatmala, Samak Kaewsuksaeng, Pongphen Jitareerat, and Masaya Kato. 2021. "Carotenoid Accumulation and the Expression of Carotenoid Metabolic Genes in Mango during Fruit Development and Ripening" Applied Sciences 11, no. 9: 4249. https://doi.org/10.3390/app11094249