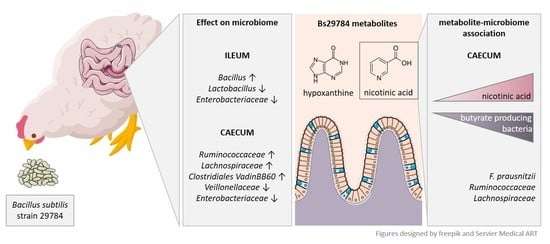
Bacillus Subtilis 29784 as a Feed Additive for Broilers Shifts the Intestinal Microbial Composition and Supports the Production of Hypoxanthine and Nicotinic Acid
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Growth Conditions
2.2. Animal Trial
2.3. Targeted Metabolomics
2.3.1. Reagents and Chemicals
2.3.2. Instrumentation
2.3.3. Optimization of the UHPLC-HRMS Method
2.3.4. Metabolomic Analysis
2.4. DNA Extraction from Intestinal Content
2.5. Quantification of Bacillus spp. and Total Bacteria
2.6. 16S rRNA Gene Amplicon Sequencing
2.7. Metabolic Function Prediction of the Microbial Communities
2.8. Statistical Analyses
3. Results
3.1. Identification of Metabolites Produced by Bs29784 In Vitro
3.2. Effect of Supplementation of Bs29784 in Broiler Feed on the Bacillus Load, Levels of Hypoxanthine and Nicotinic Acid in the Intestinal Tract
3.3. Effect of Bs29784 Supplementation in Broiler Feed on the Ileal and Cecal Microbial Diversity
3.4. Influence of Bs29784 on the Taxonomic Composition of the Ileal and Cecal Microbiome
3.5. Hypoxanthine and Nicotinic Acid Levels Are Associated with Specific Microbial Taxa in the Cecum
3.6. In-Feed Bs29784 Supplementation Decreases the Abundance of Specific Microbial Metabolic Modules
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Higgins, S.E.; Erf, G.F.; Higgins, J.P.; Henderson, S.N.; Wolfenden, A.D.; Gaona-Ramirez, G.; Hargis, B.M. Effect of probiotic treatment in broiler chicks on intestinal macrophage numbers and phagocytosis of Salmonella enteritidis by abdominal exudate cells. Poult. Sci. 2007, 86, 2315–2321. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Chen, Y.P.; Yang, M.X.; Zhang, L.L.; Lu, Z.X.; Zhou, Y.M.; Wang, T. Bacillus amyloliquefaciens supplementation alleviates immunological stress and intestinal damage in lipopolysaccharide-challenged broilers. Anim. Feed Sci. Technol. 2015, 208, 119–131. [Google Scholar] [CrossRef]
- Fan, Y.; Zhao, L.; Ji, C.; Li, X.; Jia, R.; Xi, L.; Zhang, J.; Ma, Q. Protective effects of Bacillus subtilis ANSB060 on serum biochemistry, histopathological changes and antioxidant enzyme activities of broilers fed moldy peanut meal naturally contaminated with aflatoxins. Toxins 2015, 7, 3330–3343. [Google Scholar] [CrossRef] [PubMed]
- Aliakbarpour, H.R.; Chamani, M.; Rahimi, G.; Sadeghi, A.A.; Qujeq, D. The Bacillus subtilis and lactic acid bacteria probiotics influences intestinal mucin gene expression, histomorphology and growth performance in broilers. Asian-Australas. J. Anim. Sci. 2012, 25, 1285–1293. [Google Scholar] [CrossRef] [Green Version]
- Awad, W.A.; Ghareeb, K.; Abdel-Raheem, S.; Bohm, J. Effects of dietary inclusion of probiotic and synbiotic on growth performance, organ weights, and intestinal histomorphology of broiler chickens. Poult. Sci. 2009, 88, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Bader, J.; Albin, A.; Stahl, U. Spore-forming bacteria and their utilisation as probiotics. Benef. Microbes 2012, 3, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.A.; Huang, J.M.; Khaneja, R.; Hiep, L.V.; Urdaci, M.C.; Cutting, S.M. The safety of Bacillus subtilis and Bacillus indicus as food probiotics. J. Appl. Microbiol. 2008, 105, 510–520. [Google Scholar] [CrossRef]
- Rychen, G.; Aquilina, G.; Azimonti, G.; Bampidis, V.; Bastos, M.D.L.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; Flachowsky, G.; Gropp, J.; et al. Safety and efficacy of Alterion NE® (Bacillus subtilis DSM 29784) as a feed additive for minor poultry species for fattening and reared for laying. EFSA J. 2018, 16. [Google Scholar] [CrossRef] [Green Version]
- Jacquier, V.; Nelson, A.; Jlali, M.; Rhayat, L.; Brinch, K.S.; Devillard, E. Bacillus subtilis 29,784 induces a shift in broiler gut microbiome toward butyrate-producing bacteria and improves intestinal histomorphology and animal performance. Poult. Sci. 2019, 98, 2548–2554. [Google Scholar] [CrossRef]
- Rhayat, L.; Jacquier, V.; Brinch, K.S.; Nielsen, P.; Nelson, A.; Geraert, P.A.; Devillard, E. Bacillus subtilis strain specificity affects performance improvement in broilers. Poult. Sci. 2017, 96, 2274–2280. [Google Scholar] [CrossRef]
- Neijat, M.; Shirley, R.B.; Welsher, A.; Barton, J.; Thiery, P.; Kiarie, E. Growth performance, apparent retention of components, and excreta dry matter content in Shaver White pullets (5 to 16 week of age) in response to dietary supplementation of graded levels of a single strain Bacillus subtilis probiotic. Poult. Sci. 2019, 98, 3777–3786. [Google Scholar] [CrossRef] [PubMed]
- Mohammadigheisar, M.; Shirley, R.B.; Barton, J.; Welsher, A.; Thiery, P.; Kiarie, E. Growth performance and gastrointestinal responses in heavy Tom turkeys fed antibiotic free corn−soybean meal diets supplemented with multiple doses of a single strain Bacillus subtilis probiotic (DSM29784). Poult. Sci. 2019, 98, 5541–5550. [Google Scholar] [CrossRef] [PubMed]
- Rhayat, L.; Maresca, M.; Nicoletti, C.; Perrier, J.; Brinch, K.S.; Christian, S.; Devillard, E.; Eckhardt, E. Effect of Bacillus subtilis Strains on Intestinal Barrier Function and Inflammatory Response. Front. Immunol. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.C.; Valiere, A. Probiotics and medical nutrition therapy. Nutr. Clin. Care 2004, 7, 56–68. [Google Scholar]
- Hamzehlou, P.; Sepahy, A.A.; Mehrabian, S.; Hosseini, F. Production of vitamins B3, B6 and B9 by Lactobacillus isolated from traditional yogurt samples from 3 cities in Iran, winter 2016. Appl. Food Biotechnol. 2018, 5, 105–118. [Google Scholar] [CrossRef]
- Lan, Y.; Verstegen, M.W.A.; Tamminga, S.; Williams, B.A. The role of the commensal gut microbial community in broiler chickens. Worlds. Poult. Sci. J. 2005, 61, 95–104. [Google Scholar] [CrossRef] [Green Version]
- Dickinson, D.N.; La Duc, M.T.; Haskins, W.E.; Gornushkin, I.; Winefordner, J.D.; Powell, D.H.; Venkateswaran, K. Species Differentiation of a Diverse Suite of Bacillus Spores by Mass Spectrometry-Based Protein Profiling. Appl. Environ. Microbiol. 2004, 70, 475–482. [Google Scholar] [CrossRef] [Green Version]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [Green Version]
- Rombouts, C.; Hemeryck, L.Y.; Van Hecke, T.; De Smet, S.; De Vos, W.H.; Vanhaecke, L. Untargeted metabolomics of colonic digests reveals kynurenine pathway metabolites, dityrosine and 3-dehydroxycarnitine as red versus white meat discriminating metabolites. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Vanden Bussche, J.; Marzorati, M.; Laukens, D.; Vanhaecke, L. Validated High Resolution Mass Spectrometry-Based Approach for Metabolomic Fingerprinting of the Human Gut Phenotype. Anal. Chem. 2015, 87, 10927–10934. [Google Scholar] [CrossRef]
- Kamleh, M.A.; Ebbels, T.M.D.; Spagou, K.; Masson, P.; Want, E.J. Optimizing the use of quality control samples for signal drift correction in large-scale urine metabolic profiling studies. Anal. Chem. 2012, 84, 2670–2677. [Google Scholar] [CrossRef]
- Wang, L.; Meeus, I.; Rombouts, C.; Van Meulebroek, L.; Vanhaecke, L.; Smagghe, G. Metabolomics-based biomarker discovery for bee health monitoring: A proof of concept study concerning nutritional stress in Bombus terrestris. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, R.I.; Whiteley, A.S.; O’Donnell, A.G.; Bailey, M.J. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 2000, 66, 5488–5491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguirre, M.; Vuorenmaa, J.; Valkonen, E.; Kettunen, H.; Callens, C.; Haesebrouck, F. In—Feed resin acids reduce matrix metalloproteinase activity in the ileal mucosa of healthy broilers without inducing major effects on the gut microbiota. Vet. Res. 2019, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Han, G.Q.; Xiang, Z.T.; Yu, B.; Chen, D.W.; Qi, H.W.; Mao, X.B.; Chen, H.; Mao, Q.; Huang, Z.Q. Effects of different starch sources on Bacillus spp. in intestinal tract and expression of intestinal development related genes of weanling piglets. Mol. Biol. Rep. 2012, 39, 1869–1876. [Google Scholar] [CrossRef] [Green Version]
- Hopkins, M.J.; Macfarlane, G.T.; Furrie, E.; Fite, A.; Macfarlane, S. Characterisation of intestinal bacteria in infant stools using real-time PCR and northern hybridisation analyses. FEMS Microbiol. Ecol. 2005, 54, 77–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glockner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. PANDAseq: PAired-eND Assembler for Illumina sequences. BMC Bioinform. 2012, 13, 31. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Darzi, Y.; Falony, G.; Vieira-Silva, S.; Raes, J. Towards biome-specific analysis of meta-omics data. ISME J. 2016, 10, 1025–1028. [Google Scholar] [CrossRef] [Green Version]
- Vieira-Silva, S.; Falony, G.; Darzi, Y.; Lima-Mendez, G.; Garcia Yunta, R.; Okuda, S.; Vandeputte, D.; Valles-Colomer, M.; Hildebrand, F.; Chaffron, S.; et al. Species-function relationships shape ecological properties of the human gut microbiome. Nat. Microbiol. 2016, 1, 1–8. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Dixon, P. Computer program review VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Neijat, M.; Habtewold, J.; Shirley, R.B.; Welsher, A.; Barton, J.; Thiery, P.; Kiarie, E. Bacillus subtilis Strain DSM 29784 Modulates the Cecal Microbiome, Concentration of Short-Chain Fatty Acids, and Apparent Retention of Dietary Components in Shaver White Chickens during Grower, Developer, and Laying Phases. Appl. Environ. Microbiol. 2019, 85, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Casula, G.; Cutting, S.M. Bacillus probiotics: Spore germination in the gastrointestinal tract. Appl. Environ. Microbiol. 2002, 68, 2344–2352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cartman, S.T.; La Ragione, R.M.; Woodward, M.J. Bacillus subtilis spores germinate in the chicken gastrointestinal tract. Appl. Environ. Microbiol. 2008, 74, 5254–5258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santoru, M.L.; Piras, C.; Murgia, A.; Palmas, V.; Camboni, T.; Liggi, S.; Ibba, I.; Lai, M.A.; Orrù, S.; Blois, S.; et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Soto-Martin, E.C.; Warnke, I.; Farquharson, F.M.; Christodoulou, M.; Horgan, G.; Derrien, M.; Faurie, J.M.; Flint, H.J.; Duncan, S.H.; Louis, P. Vitamin biosynthesis by human gut butyrate-producing bacteria and cross-feeding in synthetic microbial communities. MBio 2020, 11, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Magnúsdóttir, S.; Ravcheev, D.; de Crécy-Lagard, V.; Thiele, I. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front. Genet. 2015, 6, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodionov, D.A.; Arzamasov, A.A.; Khoroshkin, M.S.; Iablokov, S.N.; Leyn, S.A.; Peterson, S.N.; Novichkov, P.S.; Osterman, A.L. Micronutrient requirements and sharing capabilities of the human gut microbiome. Front. Microbiol. 2019, 10, 1316. [Google Scholar] [CrossRef] [Green Version]
- Scott Lee, J.; Wang, R.X.; Alexeev, E.E.; Lanis, J.M.; Battista, K.D.; Glover, L.E.; Colgan, S.P. Hypoxanthine is a checkpoint stress metabolite in colonic epithelial energy modulation and barrier function. J. Biol. Chem. 2018, 293, 6039–6051. [Google Scholar] [CrossRef] [Green Version]
- Santoru, M.L.; Piras, C.; Murgia, F.; Spada, M.; Tronci, L.; Leoni, V.P.; Serreli, G.; Deiana, M.; Atzori, L. Modulatory effect of nicotinic acid on the metabolism of Caco-2 cells exposed to IL-1β and LPS. Metabolites 2020, 10, 204. [Google Scholar] [CrossRef]
- Li, J.; Kong, D.; Wang, Q.; Wu, W.; Tang, Y.; Bai, T.; Guo, L.; Wei, L.; Zhang, Q.; Yu, Y.; et al. Niacin ameliorates ulcerative colitis via prostaglandin D 2 -mediated D prostanoid receptor 1 activation. EMBO Mol. Med. 2017, 9, 571–588. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Wang, R.X.; Goldberg, M.S.; Clifford, G.P.; Kao, D.J.; Colgan, S.P. Microbiota-Sourced Purines Support Wound Healing and Mucous Barrier Function. iScience 2020, 23, 101226. [Google Scholar] [CrossRef]
- Yutaka, K.; Toshiya, I.I.; Tohru, K. Absorption and metabolism of purines by the small intestine of the chicken. Comp. Biochem. Physiol. Part A Physiol. 1991, 99, 235–240. [Google Scholar] [CrossRef]
- Lee, J.S.; Wang, R.X.; Alexeev, E.E.; Colgan, S.P. Intestinal Inflammation as a Dysbiosis of Energy Procurement: New Insights into an Old Topic. Gut Microbes 2021, 13, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Galbusera, C.; Orth, P.; Fedida, D.; Spector, T. Superoxide radical production by allopurinol and xanthine oxidase. Biochem. Pharmacol. 2006, 71, 1747–1752. [Google Scholar] [CrossRef]
- Crane, J.K.; Naeher, T.M.; Broome, J.E.; Boedeker, E.C. Role of host xanthine oxidase in infection due to enteropathogenic and shiga-toxigenic Escherichia coli. Infect. Immun. 2013, 81, 1129–1139. [Google Scholar] [CrossRef] [Green Version]
- Martin, H.M.; Hancock, J.T.; Salisbury, V.; Harrison, R. Role of xanthine oxidoreductase as an antimicrobial agent. Infect. Immun. 2004, 72, 4933–4939. [Google Scholar] [CrossRef] [Green Version]
- Carro, M.D.; Falkenstein, E.; Blemings, K.P.; Klandorf, H. Determination of xanthine oxidoreductase activity in broilers: Effect of pH and temperature of the assay and distribution in tissues. Poult. Sci. 2009, 88, 2406–2414. [Google Scholar] [CrossRef]
- Graff, E.C.; Fang, H.; Wanders, D.; Judd, R.L. Anti-inflammatory effects of the hydroxycarboxylic acid receptor 2. Metabolism 2016, 65, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Ran, X.; Li, B.; Li, Y.; He, D.; Huang, B.; Fu, S.; Liu, J.; Wang, W. Sodium Butyrate Inhibits Inflammation and Maintains Epithelium Barrier Integrity in a TNBS-induced Inflammatory Bowel Disease Mice Model. EBioMedicine 2018, 30, 317–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, R.R.; Zhao, G.P.; Zhao, J.P.; Chen, J.L.; Zheng, M.Q.; Liu, R.R.; Wen, J. Influence of dietary nicotinic acid supplementation on lipid metabolism and related gene expression in two distinct broiler breeds of female chickens. J. Anim. Physiol. Anim. Nutr. 2014, 98, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Ilkhani, F.; Hosseini, B.; Saedisomeolia, A. Niacin and Oxidative Stress: A Mini-Review. J. Nutr. Med. Diet Care 2016, 2, 14. [Google Scholar] [CrossRef]
- Perumal, S.S.; Shanthi, P.; Sachdanandam, P. Augmented efficacy of tamoxifen in rat breast tumorigenesis when gavaged along with riboflavin, niacin, and CoQ10: Effects on lipid peroxidation and antioxidants in mitochondria. Chem. Biol. Interact. 2005, 152, 49–58. [Google Scholar] [CrossRef] [PubMed]







| Metabolite | Area Ratio (Mean ± SD) | Fold Change | p-Value | |
|---|---|---|---|---|
| Blank | Bs29784 | |||
| Hypoxanthine | 0.173 ± 0.002 | 1.844 ± 0.086 | 106.40 | <0.0001 |
| Nicotinic acid | 0.218 ± 0.030 | 1.853 ± 0.104 | 8.51 | <0.0001 |
| Ethanolamine | 0.007 ± 0.003 | 0.061 ± 0.016 | 8.67 | 0.005 |
| Uracil | 0.241 ± 0.004 | 1.652 ± 0.392 | 6.85 | 0.003 |
| Pantothenate | 0.001 ± 0.001 | 0.022 ± 0.002 | 2.03 | 0.002 |
| 3-Hydroxypyridine | 0.006 ± 0.003 | 0.014 ± 0.001 | 2.16 | 0.015 |
| 2.5-dimethylpyrazine | 0.005 ± 0.000 | 0.012 ± 0.003 | 2.47 | 0.017 |
| Thymine | 0.014 ± 0.007 | 0.034 ± 0.004 | 2.51 | 0.011 |
| Control | Bs29784 | p-Value | |
|---|---|---|---|
| ILEUM | |||
| Taxonomic alpha diversity | |||
| nOTUs | 98.8 ± 29.95 | 90 ± 16.02 | 0.69 |
| Chao1 | 125.31 ± 49.39 | 107.59 ± 24.07 | 0.69 |
| Shannon | 1.72 ± 0.40 | 1.06 ± 0.43 | 0.032 * |
| Functional alpha diversity | |||
| nKOs | 4487 ± 257.13 | 4522.6 ± 145.87 | 1 |
| Chao1 | 4656.89 ± 375.39 | 4743.67 ± 298.32 | 1 |
| Shannon | 7.40 ± 0.23 | 7.16 ± 0.18 | 0.15 |
| CECUM | |||
| Taxonomic alpha diversity | |||
| nOTUs | 142.8 ± 5.45 | 181.2 ± 25.08 | 0.056 |
| Chao1 | 157.74 ± 7.13 | 196.50 ± 30.77 | 0.15 |
| Shannon | 2.91 ± 0.41 | 3.26 ± 0.58 | 0.42 |
| Functional alpha diversity | |||
| nKOs | 4228.4 ± 111.10 | 4205.0 ± 76.41 | 1 |
| Chao1 | 4554.97 ± 210.53 | 4414.80 ± 191.05 | 0.42 |
| Shannon | 7.71 ± 0.13 | 7.39 ± 0.14 | 0.016 * |
| Phylum | Class | Family | Genus | Mean Abundance (%) | Log2 Fold Change | Adjusted p-Value | |
|---|---|---|---|---|---|---|---|
| Control | Bs29784 | ||||||
| ILEUM | |||||||
| Actinobacteria | Actinobacteria | Beutenbergiaceae | Ambiguous taxa Beutenbergiaceae | 0.046 | 0.000 | −23.36 | <0.001 |
| Firmicutes | Bacilli | Bacillaceae | Bacillus | 0.000 | 0.121 | 7.54 | <0.001 |
| Firmicutes | Bacilli | Lactobacillaceae | Pediococcus | 0.250 | 0.035 | −4.32 | 0.019 |
| Firmicutes | Bacilli | Leuconostocaceae | Weissella | 0.253 | 0.002 | −7.20 | <0.001 |
| Firmicutes | Clostridia | Peptostreptococcaceae | Ambiguous taxa Peptostreptococcaceae | 0.054 | 0.000 | −22.66 | <0.001 |
| Firmicutes | Negativicutes | Veillonellaceae | Family Veillonellaceae | 0.062 | 0.000 | −22.91 | <0.001 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriaceae | Ambiguous taxa Enterobacteriaceae | 0.473 | 0.051 | −3.71 | 0.007 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriaceae | Enterobacter | 0.045 | 0.002 | −6.32 | 0.001 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriaceae | Klebsiella | 0.058 | 0.002 | −6.09 | 0.007 |
| CECUM | |||||||
| Firmicutes | Bacilli | Enterococcaceae | Enterococcus | 1.746 | 4.865 | 2.30 | 0.016 |
| Firmicutes | Clostridia | Clostridiales vadinBB60 group | uncultured bacterium_Clostridiales vadinBB60 group | 0.000 | 0.956 | 12.51 | <0.001 |
| Firmicutes | Clostridia | Lachnospiraceae | [Eubacterium] hallii group | 0.000 | 0.074 | 22.48 | <0.001 |
| Firmicutes | Clostridia | Lachnospiraceae | GCA-900066575 | 0.000 | 0.062 | 22.47 | <0.001 |
| Firmicutes | Clostridia | Lachnospiraceae | Lachnospiraceae FCS020 group | 0.004 | 0.219 | 7.32 | <0.001 |
| Firmicutes | Clostridia | Lachnospiraceae | Lachnospiraceae NK4A136 group | 0.000 | 0.556 | 25.64 | <0.001 |
| Firmicutes | Clostridia | Peptostreptococcaceae | Clostridioides | 0.000 | 0.066 | 23.25 | <0.001 |
| Firmicutes | Clostridia | Ruminococcaceae | Negativibacillus | 0.000 | 0.693 | 11.10 | <0.001 |
| Firmicutes | Clostridia | Ruminococcaceae | Ruminiclostridium 9 | 0.239 | 1.359 | 2.93 | 0.0461 |
| Firmicutes | Clostridia | Ruminococcaceae | Ruminococcaceae UCG-013 | 0.000 | 0.008 | 27.52 | <0.001 |
| Firmicutes | Negativicutes | Veillonellaceae | Family_Veillonellaceae | 1.272 | 0.000 | −27.55 | <0.001 |
| Firmicutes | Negativicutes | Veillonellaceae | Sporomusa | 3.657 | 0.000 | −28.07 | <0.001 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriaceae | Ambiguous_taxa_Enterobacteriaceae | 5.518 | 0.758 | −2.48 | <0.001 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriaceae | Enterobacter | 0.718 | 0.059 | −3.03 | 0.004 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriaceae | Klebsiella | 3.221 | 0.745 | −2.33 | 0.006 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, P.; Rhayat, L.; Pinloche, E.; Devillard, E.; De Paepe, E.; Vanhaecke, L.; Haesebrouck, F.; Ducatelle, R.; Van Immerseel, F.; Goossens, E. Bacillus Subtilis 29784 as a Feed Additive for Broilers Shifts the Intestinal Microbial Composition and Supports the Production of Hypoxanthine and Nicotinic Acid. Animals 2021, 11, 1335. https://doi.org/10.3390/ani11051335
Choi P, Rhayat L, Pinloche E, Devillard E, De Paepe E, Vanhaecke L, Haesebrouck F, Ducatelle R, Van Immerseel F, Goossens E. Bacillus Subtilis 29784 as a Feed Additive for Broilers Shifts the Intestinal Microbial Composition and Supports the Production of Hypoxanthine and Nicotinic Acid. Animals. 2021; 11(5):1335. https://doi.org/10.3390/ani11051335
Chicago/Turabian StyleChoi, Pearl, Lamya Rhayat, Eric Pinloche, Estelle Devillard, Ellen De Paepe, Lynn Vanhaecke, Freddy Haesebrouck, Richard Ducatelle, Filip Van Immerseel, and Evy Goossens. 2021. "Bacillus Subtilis 29784 as a Feed Additive for Broilers Shifts the Intestinal Microbial Composition and Supports the Production of Hypoxanthine and Nicotinic Acid" Animals 11, no. 5: 1335. https://doi.org/10.3390/ani11051335