Kaposi’s Sarcoma-Associated Herpesvirus ORF7 Is Essential for Virus Production
Abstract
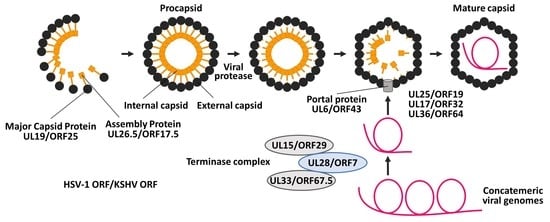
:1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Construction of ORF7-KO KSHV BAC16
2.3. Establishment of Doxycycline-Inducible Recombinant KSHV-Expressing Cells
2.4. Measurement of Virus Production and Viral DNA Replication
2.5. RT Real-Time PCR (RT-qPCR)
2.6. Complementation Assay
2.7. Antibodies
2.8. Western Blotting and Immunofluorescence Assay (IFA)
2.9. Pulldown Assays
2.10. Statistics
3. Results
3.1. Construction of Two Types of ORF7-KO KSHV BACs and Their Revertants
3.2. ORF7 Is Essential for Virus Production but Not Lytic Gene Expression and DNA Replication in KSHV
3.3. Rescue of Virus Production in iSLK-ΔORF7 Cells by ORF7 Overexpression
3.4. ORF29 and ORF67.5 Are Partially Relocalized into Nuclei from the Cytosol by ORF7 and ORF7 Colocalizes with ORF29 and ORF67.5 in the Nuclei
3.5. ORF7 Interacts with Both ORF29 and ORF67.5, Whereas ORF29 and ORF67.5 Fail to Interact with Each Other
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chang, Y.; Cesarman, E.; Pessin, M.S.; Lee, F.; Culpepper, J.; Knowles, D.M.; Moore, P.S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 1994, 266, 1865–1869. [Google Scholar] [CrossRef] [Green Version]
- Nador, R.G.; Cesarman, E.; Chadburn, A.; Dawson, D.B.; Ansari, M.Q.; Sald, J.; Knowles, D.M. Primary effusion lymphoma: A distinct clinicopathologic entity associated with the Kaposi’s sarcoma-associated herpes virus. Blood 1996, 88, 645–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soulier, J.; Grollet, L.; Oksenhendler, E.; Cacoub, P.; Cazals-Hatem, D.; Babinet, P.; d’Agay, M.F.; Clauvel, J.P.; Raphael, M.; Degos, L. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood 1995, 86, 1276–1280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russo, J.J.; Bohenzky, R.A.; Chien, M.C.; Chen, J.; Yan, M.; Maddalena, D.; Parry, J.P.; Peruzzi, D.; Edelman, I.S.; Chang, Y.; et al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 1996, 93, 14862–14867. [Google Scholar] [CrossRef] [Green Version]
- Uldrick, T.S.; Wang, V.; O’Mahony, D.; Aleman, K.; Wyvill, K.M.; Marshall, V.; Steinberg, S.M.; Pittaluga, S.; Maric, I.; Whitby, D.; et al. An interleukin-6-related systemic inflammatory syndrome in patients co-infected with Kaposi sarcoma-associated herpesvirus and HIV but without Multicentric Castleman disease. Clin. Infect. Dis. 2010, 51, 350–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, T.; Sugimoto, A.; Hosokawa, K.; Fujimuro, M. Signal transduction pathways associated with KSHV-related tumors. In Human Herpesviruses; Kawaguchi, Y., Mori, Y., Kimura, H., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; Chapter 15; pp. 321–355. [Google Scholar]
- Damania, B.; Cesarman, E. Kaposi’s sarcoma–associated herpesvirus. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 2080–2128. [Google Scholar]
- Pellett, P.E.; Roizman, B. Herpesviridae. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 1802–1822. [Google Scholar]
- Brown, J.C.; Newcomb, W.W. Herpesvirus capsid assembly: Insights from structural analysis. Curr. Opin. Virol. 2011, 1, 142–149. [Google Scholar] [CrossRef] [Green Version]
- Roizman, B.; Knipe, D.M.; Whitley, R.J. Herpes simplex viruses. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 1823–1897. [Google Scholar]
- Liu, F.Y.; Roizman, B. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J. Virol. 1991, 65, 5149–5156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newcomb, W.W.; Homa, F.L.; Thomsen, D.R.; Booy, F.P.; Trus, B.L.; Steven, A.C.; Spencer, J.V.; Brown, J.C. Assembly of the herpes simplex virus capsid: Characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 1996, 263, 432–446. [Google Scholar] [CrossRef] [Green Version]
- Trus, B.L.; Booy, F.P.; Newcomb, W.W.; Brown, J.C.; Homa, F.L.; Thomsen, D.R.; Steven, A.C. The herpes simplex virus procapsid: Structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 1996, 263, 447–462. [Google Scholar] [CrossRef]
- Addison, C.; Rixon, F.J.; Preston, V.G. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J. Gen. Virol. 1990, 71, 2377–2384. [Google Scholar] [CrossRef] [PubMed]
- Poon, A.P.; Roizman, B. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J. Virol. 1993, 67, 4497–4503. [Google Scholar] [CrossRef] [Green Version]
- Jacobson, J.G.; Yang, K.; Baines, J.D.; Homa, F.L. Linker insertion mutations in the herpes simplex virus type 1 UL28 gene: Effects on UL28 interaction with UL15 and UL33 and identification of a second-site mutation in the UL15 gene that suppresses a lethal UL28 mutation. J. Virol. 2006, 80, 12312–12323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adelman, K.; Salmon, B.; Baines, J.D. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. USA 2001, 98, 3086–3091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- al-Kobaisi, M.F.; Rixon, F.J.; McDougall, I.; Preston, V.G. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology 1991, 180, 380–388. [Google Scholar] [CrossRef]
- Yu, D.S.; Weller, K. Genetic analysis of the UL15 gene locus for the putative terminase of herpes simplex virus type 1. Virology 1998, 243, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Beard, P.M.; Taus, N.S.; Baines, J.D. DNA cleavage and packaging proteins encoded by genes UL28, UL15, and UL33 of herpes simplex virus type 1 form a complex in infected cells. J. Virol. 2002, 76, 4785–4791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.; Baines, J.D. The putative terminase subunit of herpes simplex virus 1 encoded by UL28 is necessary and sufficient to mediate interaction between pUL 15 and pUL 33. J. Virol. 2006, 80, 5733–5739. [Google Scholar] [CrossRef] [Green Version]
- Heming, J.D.; Huffman, J.B.; Jones, L.M.; Homa, F.L. Isolation and characterization of the herpes simplex virus 1 terminase complex. J. Virol. 2014, 88, 225–236. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Zhou, Z.H. Structure of the herpes simplex virus 1 capsid with associated tegument protein complexes. Science 2018, 360, 7298. [Google Scholar] [CrossRef] [Green Version]
- Tengelsen, L.A.; Pederson, N.E.; Shaver, P.R.; Wathen, M.W.; Homa, F.L. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: Isolation and characterization of two UL28 deletion mutants. J. Virol. 1993, 67, 3470–3480. [Google Scholar] [CrossRef] [Green Version]
- Tsurumi, S.; Watanabe, T.; Iwaisako, Y.; Suzuki, Y.; Nakano, T.; Fujimuro, M. Kaposi’s sarcoma-associated herpesvirus ORF17 plays a key role in capsid maturation. Virology 2021, 558, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Tischer, B.K.; Smith, G.A.; Osterrieder, N. En passant mutagenesis: A two step markerless red recombination system. Methods Mol. Biol. 2010, 634, 421–430. [Google Scholar] [PubMed]
- Brulois, K.F.; Chang, H.; Lee, A.S.; Ensser, A.; Wong, L.Y.; Toth, Z.; Lee, S.H.; Lee, H.R.; Myoung, J.; Ganem, D.; et al. Construction and manipulation of a new Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome clone. J. Virol. 2012, 86, 9708–9720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, T.; Nishimura, M.; Izumi, T.; Kuriyama, K.; Iwaisako, Y.; Hosokawa, K.; Takaori-Kondo, A.; Fujimuro, M. Kaposi’s Sarcoma-Associated Herpesvirus ORF66 Is Essential for Late Gene Expression and Virus Production via Interaction with ORF34. J. Virol. 2020, 94, e01300-19. [Google Scholar] [CrossRef]
- Nishimura, M.; Watanabe, T.; Yagi, S.; Yamanaka, T.; Fujimuro, M. Kaposi’s sarcoma-associated herpesvirus ORF34 is essential for late gene expression and virus production. Sci. Rep. 2017, 7, 329. [Google Scholar] [CrossRef] [Green Version]
- Arias, C.; Weisburd, B.; Stern-Ginossar, N.; Mercier, A.; Madrid, A.S.; Bellare, P.; Holdorf, M.; Weissman, J.S.; Ganem, D. KSHV 2.0: A comprehensive annotation of the Kaposi’s sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog. 2014, 10, e1003847. [Google Scholar] [CrossRef]
- Gong, D.; Dai, X.; Jih, J.; Liu, Y.T.; Bi, G.Q.; Sun, R.; Zhou, Z.H. DNA-Packing Portal and Capsid-Associated Tegument Complexes in the Tumor Herpesvirus KSHV. Cell 2019, 178, 1329–1343. [Google Scholar] [CrossRef] [Green Version]
- Gao, M.; Matusick-Kumar, L.; Hurlburt, W.; DiTusa, S.F.; Newcomb, W.W.; Brown, J.C.; McCann, P.J., 3rd; Deckman, I.; Colonno, R.J. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J. Virol. 1994, 68, 3702–3712. [Google Scholar] [CrossRef] [Green Version]
- Dunn, W.; Chou, C.; Li, H.; Hai, R.; Patterson, D.; Stolc, V.; Zhu, H.; Liu, F. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 2003, 100, 14223–14228. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.; Silva, M.C.; Shenk, T. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 2003, 100, 12396–12401. [Google Scholar] [CrossRef] [Green Version]
- Chiu, S.H.; Wu, M.C.; Wu, C.C.; Chen, Y.C.; Lin, S.F.; Hsu, J.T.; Yang, C.S.; Tsai, C.H.; Takada, K.; Chen, M.R.; et al. Epstein-Barr virus BALF3 has nuclease activity and mediates mature virion production during the lytic cycle. J. Virol. 2014, 88, 4962–4975. [Google Scholar] [CrossRef] [Green Version]
- Goldner, T.; Hewlett, G.; Ettischer, N.; Ruebsamen-Schaeff, H.; Zimmermann, H.; Lischka, P. The novel anticytomegalovirus compound AIC246 (Letermovir) inhibits human cytomegalovirus replication through a specific antiviral mechanism that involves the viral terminase. J. Virol. 2011, 85, 10884–10893. [Google Scholar] [CrossRef] [Green Version]
- Baines, J.D.; Cunningham, C.; Nalwanga, D.; Davison, A. The U(L)15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the U(L)15 gene product. J. Virol. 1997, 71, 2666–2673. [Google Scholar] [CrossRef] [Green Version]
- Koslowski, K.M.; Shaver, P.R.; Casey, J.T., 2nd; Wilson, T.; Yamanaka, G.; Sheaffer, A.K.; Tenney, D.J.; Pederson, N.E. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J. Virol. 1999, 73, 1704–1707. [Google Scholar] [CrossRef] [Green Version]
- Higgs, M.R.; Preston, V.G.; Stow, N.D. The UL15 protein of herpes simplex virus type 1 is necessary for the localization of the UL28 and UL33 proteins to viral DNA replication centres. J. Gen. Virol. 2008, 89, 1709–1715. [Google Scholar] [CrossRef]
- Yang, K.; Dang, X.; Baines, J.D. A Domain of Herpes Simplex Virus pUL33 Required to Release Monomeric Viral Genomes from Cleaved Concatemeric DNA. J. Virol. 2017, 91, e00854-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Common Name | HSV-1 | KSHV |
|---|---|---|
| capsid maturation protease | UL26 (VP24, VP21) | ORF17 |
| assembly protein | UL26.5 | ORF17.5 |
| major capsid protein | UL19 | ORF25 |
| triplex protein 1 | UL38 | ORF62 |
| triplex protein 2 | UL18 | ORF26 |
| small capsid protein | UL35 | ORF65 |
| DNA packaging terminase subunit 1 | UL15 | ORF29 |
| DNA packaging terminase subunit 2 | UL28 | ORF7 |
| DNA packaging protein | UL33 | ORF67.5 |
| portal protein | UL6 | ORF43 |
| portal capping protein | UL25 | ORF19 |
| DNA packaging tegument protein | UL17 | ORF32 |
| large tegument protein | UL36 | ORF64 |
| Primer Name | Primer Sequences (5′ -> 3′) |
|---|---|
| BAC Mutagenesis a | |
| S_dORF7_fs_EP | ctctctgacctggatttgtagttgtgtacccgtaacgatg*caaaggaactggcggcggtcTAGGGATAACAGGGTAATCGATTT |
| As_dORF7_fs_EP | agggctgacacatcggcatagaccgccgccagttcctttg*catcgttacgggtacacaacGCCAGTGTTACAACCAATTAACC |
| S_dORF7_st_EP | gtgtctgactcccaaccagggcaccagtctgcaggccatgTAGTTAGATAGTctcccagacacggcctgcagTAGGGATAACAGGGTAATCGATTT |
| As_dORF7_st_EP | atgcgggggtacatatgtgactgcaggccgtgtctgggagACTATCTAACTAcatggcctgcagactggtgcGCCAGTGTTACAACCAATTAACC |
| S_dORF7_fs-rev_EP | ctctctgacctggatttgtagttgtgtacccgtaacgatggcaaaggaactggcggcggtcTAGGGATAACAGGGTAATCGATTT |
| As_dORF7_fs-rev_EP | agggctgacacatcggcatagaccgccgccagttcctttgccatcgttacgggtacacaacGCCAGTGTTACAACCAATTAACC |
| S_dORF7_st-rev_EP | gtgtctgactcccaaccagggcaccagtctgcaggccatgctcccagacacggcctgcagTAGGGATAACAGGGTAATCGATTT |
| As_dORF7_st-rev_EP | atgcgggggtacatatgtgactgcaggccgtgtctgggagcatggcctgcagactggtgcGCCAGTGTTACAACCAATTAACC |
| Expression Plasmid b | |
| S_EcoRI_ORF7 | catGAATTCatggcaaaggaactggcggc |
| As_XbaI_ORF7 | gtcTCTAGAgacctgggagtcattgtggttgc |
| S_EcoRI_ORF17 | catGAATTCatggcacagggcctgtacgtc |
| As_SalI_ORF17 | tagGTCGACctactgcttgttcaggagctc |
| S_EcoRI_ORF29 | catGAATTCatgctgctcagccgtcacag |
| As_SalI_ORF29 | taaGTCGACttattgtggggatatgggcttgtac |
| S_EcoRI_ORF67.5 | catGAATTCatggagtacgcgtctgaccag |
| As_SalI_ORF67.5 | tgaGTCGACtcagggccgtgc |
| Real-Time PCR | |
| KSHV ORF11-F | TTGACAACACGCACCGCAAG |
| KSHV ORF11-R | AAAAATCAGCACGCTCGAGGAG |
| RT Real-Time PCR | |
| Fw-GAPDH | CATCAAGAAGGTGGTGAAGCAG |
| Rv-GAPDH | TGTCGCTGTTGAAGTCAGAGG |
| Fw-ORF16 | AGATTTCACAGCACCACCGGTA |
| Rv-ORF16 | CCCCAGTTCATGTTTCCATCGC |
| Fw-ORF47 | CGATCCGAATCACTGCAACG |
| Rv-ORF47 | CTGCTGCTTTTAGCCCGAG |
| Fw-K8.1 | TCCCACGTATCGTTCGCATTTGG |
| Rv-K8.1 | GCGTCTCTTCCTCTAGTCGTTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iwaisako, Y.; Watanabe, T.; Hanajiri, M.; Sekine, Y.; Fujimuro, M. Kaposi’s Sarcoma-Associated Herpesvirus ORF7 Is Essential for Virus Production. Microorganisms 2021, 9, 1169. https://doi.org/10.3390/microorganisms9061169
Iwaisako Y, Watanabe T, Hanajiri M, Sekine Y, Fujimuro M. Kaposi’s Sarcoma-Associated Herpesvirus ORF7 Is Essential for Virus Production. Microorganisms. 2021; 9(6):1169. https://doi.org/10.3390/microorganisms9061169
Chicago/Turabian StyleIwaisako, Yuki, Tadashi Watanabe, Mizuki Hanajiri, Yuichi Sekine, and Masahiro Fujimuro. 2021. "Kaposi’s Sarcoma-Associated Herpesvirus ORF7 Is Essential for Virus Production" Microorganisms 9, no. 6: 1169. https://doi.org/10.3390/microorganisms9061169