Isolation and Characterization of Novel Bacteria Capable of Degrading 1,4-Dioxane in the Presence of Diverse Co-Occurring Compounds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
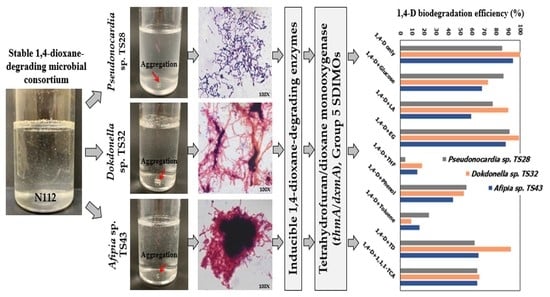
2.2. Isolation and Identification of Metabolic 1,4-D-Degrading Bacteria
2.3. Analytical Procedures
2.4. Preparation of Inocula of Isolated Strains for 1,4-D Degradation Experiments
2.5. Evaluation of 1,4-D-Degrading Enzyme Inducibility
2.6. Detection and Localization of Involed SDIMO Genes
2.7. Evaluation of Substrate Range
2.8. 1,4-D Degradation Experiments in the Presence of Additional Carbon Sources
3. Results
3.1. Isolation and Identification of Metabolic 1,4-D-Degrading Bacteria
3.2. Inducibility of 1,4-D-Degrading Enzymes
3.3. Detection and Localization of SDIMO Genes in the Isolated Strains
3.4. Substrate Range of the Isolated Strains
3.5. 1,4-D Degradation Efficiency of the Isolated Strains in the Presence of Additional Carbon Source
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Inoue, D.; Hisada, K.; Okumura, T.; Yabuki, Y.; Yoshida, G.; Kuroda, M.; Ike, M. Carbon sources that enable enrichment of 1,4-dioxane-degrading bacteria in landfill leachate. Biodegradation 2019, 31, 23–34. [Google Scholar] [CrossRef]
- Duncan, B.; Vavricka, E.; Morrison, R. A forensic overview of 1,4-dioxane. Environ. Claims J. 2004, 16, 69–79. [Google Scholar] [CrossRef]
- Zenker, M.J.; Borden, R.C.; Barlaz, M.A. Occurrence and treatment of 1,4-dioxane in aqueous environments. Environ. Eng. Sci. 2003, 20, 423–432. [Google Scholar] [CrossRef]
- Han, J.S.; So, M.H.; Kim, C.G. Optimization of biological wastewater treatment conditions for 1,4-dioxane decomposition in polyester manufacturing processes. Water Sci. Technol. 2009, 59, 995–1002. [Google Scholar] [CrossRef]
- Barndōk, H.; Cortijo, L.; Hermosilla, D.; Negro, C.; Blanco, Á. Removal of 1,4-dioxane from industrial wastewaters: Routes of decomposition under different operational conditions to determine the ozone oxidation capacity. J. Hazard. Mater. 2014, 280, 340–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Gedalanga, P.B.; Mahendra, S. Advances in bioremediation of 1,4-dioxane-contaminated waters. J. Environ. Manag. 2017, 24, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Stepien, D.K.; Diehl, P.; Helm, J.; Thomas, A.; Püttmann, W. Fate of 1,4-dioxane in the aquatic environment: From sewage to drinking water. Water Res. 2014, 48, 406–419. [Google Scholar] [CrossRef] [PubMed]
- Adamson, D.T.; Mahendra, S.; Walker, K.L., Jr.; Rauch, S.R.; Sengupta, S.; Newell, C.J. A multisite survey to identify the scale of the 1,4-dioxane problem at contaminated groundwater sites. Environ. Sci. Technol. Lett. 2014, 1, 254–258. [Google Scholar] [CrossRef]
- Adamson, D.T.; de Blanc, P.C.; Farhat, S.K.; Newell, C.J. Implications of matrix diffusion on 1,4-dioxane persistence at contaminated groundwater sites. Sci. Total Environ. 2016, 562, 98–107. [Google Scholar] [CrossRef]
- IARC (International Agency for Research on Cancer). Re-evaluation of some organic chemicals, hydrazine and hydrogen peroxide. In IARC Monographs on the Evaluation of Carcinogenic Risks to Humans; No. 71; IARC: Lyon, France, 1999. Available online: https://www.ncbi.nlm.nih.gov/books/NBK498701/ (accessed on 21 March 2021).
- WHO (World Health Organization). Guidelines for Drinking-Water Quality, 3rd ed.; Recommendations; WHO: Geneva, Switzerland, 1993; Volume 1, Available online: https://www.who.int/water_sanitation_health/dwq/GDWQ2004web.pdf?ua=1 (accessed on 21 March 2021).
- USEPA (U.S. Environmental Protection Agency). Statistics for the New Chemicals Review Program under TSCA; USEPA: Washington, DC, USA, 2016. Available online: https://www.epa.gov/reviewing-new-chemicals-under-toxic-substances-control-act-tsca/statistics-new-chemicals-review (accessed on 21 March 2021).
- Yamamoto, N.; Saito, Y.; Inoue, D.; Sei, K.; Ike, M. Characterization of newly isolated Pseudonocardia sp. N23 with high 1,4-dioxane-degrading ability. J. Biosci. Bioeng. 2018, 125, 552–558. [Google Scholar] [CrossRef]
- Deng, D.; Li, F.; Wu, C.; Li, M. Synchronic biotransformation of 1,4-dioxane and 1,1-dichloroethylene by a gram-negative propanotroph Azoarcus sp. DD4. Environ. Sci. Technol. Lett. 2018, 5, 526–532. [Google Scholar] [CrossRef]
- Tusher, T.R.; Shimizu, T.; Inoue, C.; Chien, M.-F. Enrichment and analysis of stable 1,4-dioxane-degrading microbial consortia consisting of novel dioxane-degraders. Microorganisms 2020, 8, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, F.; Wang, Y.; Yang, J.; Guo, H.; Su, D.; Yu, L. Degradation of 1,4-dioxane by Xanthobacter sp. YN2. Curr. Microbiol. 2021, 78, 992–1005. [Google Scholar] [CrossRef]
- Kim, C.G.; Seo, H.J.; Lee, B.R. Decomposition of 1,4-dioxane by advanced oxidation and biochemical process. J. Environ. Sci. Health A Toxic Hazard. Subst. Environ. Eng. 2006, 41, 599–611. [Google Scholar] [CrossRef]
- Mahendra, S.; Alvarez-Cohen, L. Kinetics of 1,4-dioxane biodegradation by monooxygenase-expressing bacteria. Environ. Sci. Technol. 2006, 40, 5435–5442. [Google Scholar] [CrossRef] [PubMed]
- Gedalanga, P.B.; Pornwongthong, P.; Mora, R.; Chiang, S.-Y.D.; Baldwin, B.; Ogles, D.; Mahendra, S. Identification of biomarker genes to predict biodegradation of 1,4-dioxane. Appl. Environ. Microbiol. 2014, 80, 3209–3218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coleman, N.V.; Bui, N.B.; Holmes, A.J. Soluble di-iron monooxygenase gene diversity in soils, sediments and ethene enrichments. Environ. Microbiol. 2006, 8, 1228–1239. [Google Scholar] [CrossRef]
- Leahy, J.G.; Batchelor, P.J.; Morcomb, S.M. Evolution of the soluble diiron monooxygenases. FEMS Microbiol. Rev. 2006, 27, 449–479. [Google Scholar] [CrossRef]
- Li, F.; Deng, D.; Li, M. Distinct catalytic behaviors between two 1,4-dioxane-degrading monooxygenases: Kinetics, inhibition, and substrate range. Environ. Sci. Technol. 2020, 54, 1898–1908. [Google Scholar] [CrossRef]
- Sun, B.; Ko, K.; Ramsay, J.A. Biodegradation of 1,4-dioxane by a Flavobacterium. Biodegradation 2011, 22, 651–659. [Google Scholar] [CrossRef]
- Rolston, H.M.; Hyman, M.R.; Semprini, L. Aerobic cometabolism of 1,4-dioxane by isobutene-utilizing microorganisms including Rhodococcus rhodochrous strain 21198 in aquifer microcosms: Experimental and modeling study. Sci. Total Environ. 2019, 694, 133688. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Wie, Y.M.; Jahng, D.; Yeom, I.T. Effects of additional carbon sources in the biodegradation of 1,4-dioxane by a mixed culture. Water 2020, 12, 1718. [Google Scholar] [CrossRef]
- Inoue, D.; Tsunoda, T.; Yamamoto, N.; Ike, M.; Sei, K. 1,4-dioxane degradation characteristics of Rhodococcus aetherivorans JCM 14343. Biodegradation 2018, 29, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Huang, H.; Shen, D. Multi-substrate biodegradation interaction of 1,4-dioxane and BTEX mixtures by Acinetobacter baumannii DD1. Biodegradation 2016, 27, 37–46. [Google Scholar] [CrossRef]
- Pugazhendi, A.; Banu, J.R.; Dhavamani, J.; Yeom, I.T. Biodegradation of 1,4-dioxane by Rhodanobacter AYS5 and the role of additional substrates. Ann. Microbiol. 2015, 65, 2201–2208. [Google Scholar] [CrossRef]
- Zhang, S.; Gedalanga, P.B.; Mahendra, S. Biodegradation kinetics of 1,4-dioxane in chlorinated solvent mixtures. Environ. Sci. Technol. 2016, 50, 9599–9607. [Google Scholar] [CrossRef]
- Chen, D.-Z.; Jin, X.-J.; Chen, J.; Ye, J.-X.; Jiang, N.-X.; Chen, J.-M. Intermediates and substrate interaction of 1,4-dioxane degradation by the effective metabolizer Xanthobacter flavus DT8. Int. Biodeter. Biodegr. 2016, 106, 133–140. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Li, M.; Conlon, P.; Fiorenza, S.; Vitale, R.J.; Alvarez, P.J.J. Rapid analysis of 1,4-dioxane in groundwater by frozen micro-extraction with gas chromatography/mass spectrometry. Groundwater Monit. Rem. 2011, 31, 70–76. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Grostern, A.; Sales, C.M.; Zhuang, W.-Q.; Erbilgin, O.; Alvarez-Cohen, L. Glyoxylate metabolism is a key feature of the metabolic degradation of 1,4-dioxane by Pseudonocardia dioxanivorans strain CB1190. Appl. Environ. Microbiol. 2012, 78, 3298–3308. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Mason, O.U.; Lowe, A.; Zhou, C.; Chen, G.; Tang, Y. Microbial community analysis provides insights into the effects of tetrahydrofuran on 1,4-dioxane biodegradation. Appl. Environ. Microbiol. 2019, 85, e00244–e19. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Mason, O.U.; Lowe, A.; Zhang, Z.; Zhou, C.; Chen, G.; Villalonga, M.J.; Tang, Y. Investigating promising substrates for promoting 1,4-dioxane biodegradation: Effects of ethane and tetrahydrofuran on microbial consortia. Biodegradation 2020, 31, 171–182. [Google Scholar] [CrossRef]
- Inoue, D.; Yoshikawa, T.; Okumura, T.; Yabuki, Y.; Ike, M. Treatment of 1,4-dioxane-containing water using carriers immobilized with indigenous microorganisms in landfill leachate treatment sludge: A laboratory-scale reactor study. J. Hazard. Mater. 2021, 414, 125497. [Google Scholar] [CrossRef]
- Sei, K.; Miyagaki, K.; Kakinoki, T.; Fukugasako, K.; Inoue, D.; Ike, M. Isolation and characterization of bacterial strains that have high ability to degrade 1,4-dioxane as a sole carbon and energy source. Biodegradation 2013, 24, 665–674. [Google Scholar] [CrossRef]
- Huang, H.; Shen, D.; Li, N.; Shan, D.; Shentu, J.; Zhou, Y. Biodegradation of 1,4-dioxane by a novel strain and its biodegradation pathway. Water Air Soil Pollut. 2014, 225, 2135. [Google Scholar] [CrossRef]
- Jin, X.J.; Chen, D.Z.; Zhu, R.Y.; Chen, J.; Chen, J.M. Characteristics of 1,4-dioxane degradation by Xanthobacter flavus DT8. Environ. Sci. 2012, 33, 1657–1662. (In Chinese) [Google Scholar]
- Nam, J.-H.; Ventura, J.-R.S.; Yeom, I.T.; Lee, Y.; Jahng, D. Structural and kinetic characteristics of 1,4-dioxane-degrading bacterial consortia containing the phylum TM7. J. Microbiol. Biotechnol. 2016, 26, 1951–1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, X.; Liu, F.; Wang, J.; Li, C.; Zheng, X. Mechanism of 1,4-dioxane microbial degradation revealed by 16S rRNA and metatranscriptomic analyses. Water Sci. Technol. 2018, 77, 123–133. [Google Scholar] [CrossRef]
- Inoue, D.; Tsunoda, T.; Sawada, K.; Yamamoto, N.; Saito, Y.; Sei, K.; Ike, M. 1,4-dioxane degradation potential of members of the genera Pseudonocardia and Rhodococcus. Biodegradation 2016, 27, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Masuda, H.; McClay, K.; Steffan, R.J.; Zylstra, G.J. Biodegradation of tetrahydrofuran and 1,4-dioxane by soluble diiron monooxygenase in Pseudonocardia sp. strain ENV478. J. Mol. Microbiol. Biotechnol. 2012, 22, 312–316. [Google Scholar] [CrossRef]
- Sales, C.M.; Grostern, A.; Parales, J.V.; Parales, R.E.; Alvarez-Cohen, L. Oxidation of the cyclic ethers 1,4-dioxane and tetrahydrofuran by a monooxygenase in two Pseudonocardia species. Appl. Environ. Microbiol. 2013, 79, 7702–7708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Mathieu, J.; Liu, Y.; Van Orden, E.T.; Yang, Y.; Fiorenza, S.; Alvarez, P.J.J. The abundance of tetrahydrofuran/dioxane monooxygenase genes (thmA/dxmA) and 1,4-dioxane degradation activity are significantly correlated at various impacted aquifers. Environ. Sci. Technol. Lett. 2014, 1, 122–127. [Google Scholar] [CrossRef]
- He, Y.; Mathieu, J.; Yang, Y.; Yu, P.; Silva, M.L.B.D.; Alvarez, P.J.J. 1,4-dioxane biodegradation by Mycobacterium dioxanotrophicus PH-06 is associated with a group-6 soluble di-iron monooxygenase. Environ. Sci. Technol. Lett. 2017, 4, 494–499. [Google Scholar] [CrossRef] [Green Version]
- Sales, C.M.; Mahendra, S.; Grostern, A.; Parales, R.E.; Goodwin, L.A.; Woyke, T.; Nolan, M.; Lapidus, A.; Chertkov, O.; Ovchinnikova, G. Genome sequence of the 1,4-dioxane-degrading Pseudonocardia dioxanivorans strain CB1190. J. Bacteriol. 2011, 193, 4549–4550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basak, B.; Chakraborty, S.; Bhunia, B.; Dey, A. Microbial remediation of recalcitrant aromatic compounds. In Industrial & Environmental Biotechnology; Pramanik, K., Patra, J.K., Eds.; Studium Press (India) Pvt. Ltd.: New Delhi, India, 2014; pp. 172–189. [Google Scholar]
- Li, M.; Liu, Y.; He, Y.; Mathieu, J.; Hatton, J.; DiGuiseppi, W.; Alvarez, P.J.J. Hindrance of 1,4-dioxane biodegradation in microcosms biostimulated with inducing or non-inducing auxiliary substrates. Water Res. 2017, 112, 217–225. [Google Scholar] [CrossRef]
- Nahar, N.; Alauddin, M.; Quilty, B. Toxic effects of toluene on the growth of activated sludge bacteria. World J. Microbiol. Biotechnol. 2000, 16, 307–311. [Google Scholar] [CrossRef]





| Substrates | 1,4-D Degradation Efficiency (%) | ||
|---|---|---|---|
| Strain TS28 | Strain TS32 | Strain TS43 | |
| 1,4-D only | 85.3 ± 5.66 | 100.0 ± 0.00 | 94.6 ± 7.60 |
| 1,4-D + glucose | 85.7 ± 0.02 | 72.6 ± 0.13 | 67.6 ± 2.34 |
| 1,4-D + LA | 77.6 ± 12.69 | 90.0 ± 14.16 | 59.0 ± 0.45 |
| 1,4-D + EG | 90.8 ± 6.41 | 99.0 ± 0.25 | 88.0 ± 1.75 |
| 1,4-D + THF | 4.4 ± 0.94 | 18.5 ± 12.11 | 14.0 ± 5.33 |
| 1,4-D + phenol | 55.4 ± 1.70 | 53.0 ± 1.60 | 43.7 ± 3.31 |
| 1,4-D + TD | 62.5 ± 16.91 | 92.3 ± 4.43 | 65.1 ± 3.04 |
| 1,4-D + toluene | 24.0 ± 0.54 | 8.7 ± 5.58 | 15.7 ± 4.76 |
| 1,4-D + 1,1,1-TCA | 63.9 ± 0.36 | 66.5 ± 2.50 | 63.8 ± 6.38 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tusher, T.R.; Shimizu, T.; Inoue, C.; Chien, M.-F. Isolation and Characterization of Novel Bacteria Capable of Degrading 1,4-Dioxane in the Presence of Diverse Co-Occurring Compounds. Microorganisms 2021, 9, 887. https://doi.org/10.3390/microorganisms9050887
Tusher TR, Shimizu T, Inoue C, Chien M-F. Isolation and Characterization of Novel Bacteria Capable of Degrading 1,4-Dioxane in the Presence of Diverse Co-Occurring Compounds. Microorganisms. 2021; 9(5):887. https://doi.org/10.3390/microorganisms9050887
Chicago/Turabian StyleTusher, Tanmoy Roy, Takuya Shimizu, Chihiro Inoue, and Mei-Fang Chien. 2021. "Isolation and Characterization of Novel Bacteria Capable of Degrading 1,4-Dioxane in the Presence of Diverse Co-Occurring Compounds" Microorganisms 9, no. 5: 887. https://doi.org/10.3390/microorganisms9050887