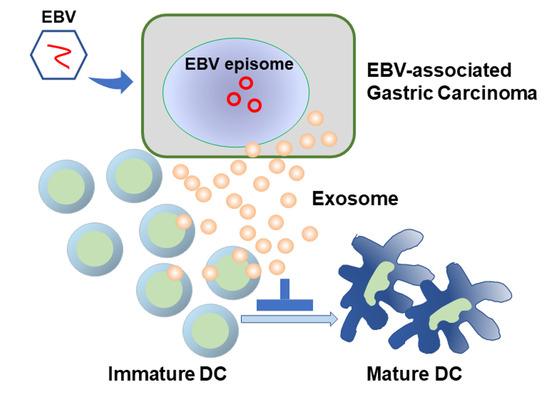
Exosomes of Epstein-Barr Virus-Associated Gastric Carcinoma Suppress Dendritic Cell Maturation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture of Gastric Cancer Cell Lines
2.2. RNA Isolation and RT-PCR
2.3. Exosome Isolation and Purification
2.4. Preparation of Whole Cell Lysates
2.5. Western Blotting
2.6. Transmission Electron Microscopy
2.7. Culture and Stimulation of Dendritic Cells Derived from Human Monocytes
2.8. Tissue Samples
2.9. Immunohistochemistry
2.10. Analysis of Correlation between the Density of Dendritic Cells and Clinicopathological Characteristics
2.11. Statistical Analysis
3. Results
3.1. Exosomes Purified from Gastric Cancer Cell Lines with and without EBV Infection
3.2. Effect of Artificial EBV-Infection on Exosomes of Gastric Cancer Cells
3.3. Exosomes of EBVaGC Gastric Cancer Cells
3.4. FACS Analysis of Human Monocyte-Derived Dendritic Cells Incubated with Gastric Cancer Exosomes
3.5. Immunohistochemical Detection of Dendritic Cells in Gastric Cancer Tissue
3.6. Correlation between Density of Dendritic Cells and Clinicopathological Factors
3.7. Assessment of Mature DC Ratio in DC-High and DC-Low EBVaGC Cases
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Estimating the Number of Exosomes
References
- Young, L.S.; Rickinson, A.B. Epstein–Barr virus: 40 years on. Nat. Rev. Cancer 2004, 4, 757–768. [Google Scholar] [CrossRef]
- Fukayama, M.; Abe, H.; Kunita, A.; Shinozaki-Ushiku, A.; Matsusaka, K.; Ushiku, T.; Kaneda, A. Thirty years of Epstein-Barr virus-associated gastric carcinoma. Virchows Arch. 2020, 476, 353–365. [Google Scholar] [CrossRef]
- Sano, T.; Coit, D.G.; Kim, H.H.; Roviello, F.; Kassab, P.; Wittekind, C.; Yamamoto, Y.; Ohashi, Y. Proposal of a new stage grouping of gastric cancer for TNM classification: International Gastric Cancer Association staging project. Gastric Cancer 2017, 20, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Balkwill, F.R.; Capasso, M.; Hagemann, T. The tumor microenvironment at a glance. J. Cell Sci. 2012, 125, 5591–5596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Yu, D. Exosomes in cancer development, metastasis, and immunity. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, M.D.; Cosmopoulos, K.; Thorley-Lawson, D.A.; van Eijndhoven, M.A.; Hopmans, E.S.; Lindenberg, J.L.; de Gruijl, T.D.; Würdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saiki, Y.; Ohtani, H.; Naito, Y.; Miyazawa, M.; Nagura, H. Immunophenotypic characterization of Epstein-Barr virus-associated gastric carcinoma: Massive infiltration by proliferating CD8+ T-lymphocytes. Lab. Invest. 1996, 75, 67–76. [Google Scholar] [PubMed]
- Kijima, Y.; Ishigami, S.; Hokita, S.; Koriyama, C.; Akiba, S.; Eizuru, Y.; Aikou, T. The comparison of the prognosis between Epstein-Barr virus (EBV)-positive gastric carcinomas and EBV-negative ones. Cancer Lett. 2003, 200, 33–40. [Google Scholar] [CrossRef]
- van Beek, J.; zur Hausen, A.; Snel, S.N.; Berkhof, J.; Kranenbarg, E.; van de Velde, C.J.; van den Brule, A.J.; Middeldorp, J.M.; Meijer, C.J.; Bloemena, E. Morphological evidence of an activated cytotoxic T-cell infiltrate in EBV-positive gastric carcinoma preventing lymph node metastases. Am. J. Surg. Pathol. 2006, 30, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.P.; Chen, J.N.; Xiao, L.; He, Q.; Feng, Z.Y.; Zhang, Z.G.; Liu, J.P.; Wei, H.B.; Shao, C.K. The implication of tumor-infiltrating lymphocytes in Epstein-Barr virus-associated gastric carcinoma. Hum. Pathol. 2019, 85, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Hubert, M.; Gobbini, E.; Bendriss-Vermare, N.; Caux, C.; Valladeau-Guilemond, J. Human tumor-infiltrating dendritic cells: From in situ visualization to high-dimensional analyses. Cancers 2019, 11, 1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Surówka, J.; Wertel, I.; Okła, K.; Bednarek, W.; Tarkowski, R.; Kotarski, J. Influence of ovarian cancer type I and type II microenvironment on the phenotype and function of monocyte-derived dendritic cells. Clin. Transl. Oncol. 2017, 19, 1489–1497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imai, S.; Nishikawa, J.; Takada, K. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J. Virol. 1998, 72, 4371–4378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshioka, Y.; Konishi, Y.; Kosaka, N.; Katsuda, T.; Kato, T.; Ochiya, T. Comparative marker analysis of extracellular vesicles in different human cancer types. J. Extracell. Vesicles 2013, 18, 2. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org/ (accessed on 25 September 2020).
- Nakayama, A.; Abe, H.; Kunita, A.; Saito, R.; Kanda, T.; Yamashita, H.; Seto, Y.; Ishikawa, S.; Fukayama, M. Viral loads correlate with upregulation of PD-L1 and worse patient prognosis in Epstein-Barr Virus-associated gastric carcinoma. PLoS ONE 2019, 14, e0211358. [Google Scholar] [CrossRef]
- Zhao, M.; Nanbo, A.; Sun, L.; Lin, Z. Extracellular vesicles in Epstein-Barr virus’ life cycle and pathogenesis. Microorganisms 2019, 7, 48. [Google Scholar] [CrossRef] [Green Version]
- Liao, C.; Zhou, Q.; Zhang, Z.; Wu, X.; Zhou, Z.; Li, B.; Peng, J.; Shen, L.; Li, D.; Luo, X.; et al. Epstein-Barr virus-encoded latent membrane protein 1 promotes extracellular vesicle secretion through syndecan-2 and synaptotagmin-like-4 in nasopharyngeal carcinoma cells. Cancer Sci. 2020, 111, 857–868. [Google Scholar] [CrossRef]
- Lee, C.P.; Liu, P.T.; Kung, H.N.; Su, M.T.; Chua, H.H.; Chang, Y.H.; Chang, C.W.; Tsai, C.H.; Liu, F.T.; Chen, M.R. The ESCRT machinery is recruited by the viral BFRF1 protein to the nucleus-associated membrane for the maturation of Epstein-Barr Virus. PLoS Pathog. 2012, 8, e1002904. [Google Scholar] [CrossRef] [Green Version]
- Braz-Silva, P.H.; Vitale, S.; Butori, C.; Guevara, N.; Santini, J.; Magalhães, M.; Hofman, P.; Doglio, A. Specific infiltration of langerin-positive dendritic cells in EBV-infected tonsil, Hodgkin lymphoma and nasopharyngeal carcinoma. Int. J. Cancer 2011, 128, 2501–2508. [Google Scholar] [CrossRef]
- Nilsson, J.S.; Abolhalaj, M.; Lundberg, K.; Lindstedt, M.; Greiff, L. Dendritic cell subpopulations in nasopharyngeal cancer. Oncol. Lett. 2019, 17, 2557–2561. [Google Scholar] [CrossRef] [Green Version]
- Manz, M.G. Plasmacytoid dendritic cells: Origin matters. Nat. Immunol. 2018, 19, 652–654. [Google Scholar] [CrossRef] [PubMed]
- Borozan, I.; Zapatka, M.; Frappier, L.; Ferretti, V. Analysis of Epstein-Barr Virus Genomes and Expression Profiles in Gastric Adenocarcinoma. J. Virol. 2018, 92, e01239-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanbo, A.; Ohashi, M.; Yoshiyama, H.; Ohba, Y. The Role of Transforming Growth Factor β in Cell-to-Cell Contact-Mediated Epstein-Barr Virus Transmission. Front Microbiol. 2018, 9, 984. [Google Scholar] [CrossRef] [PubMed]







| Protein (µg/105 Cell) Fold | CD63 Density Fold | CD81 Density Fold | Size | |
|---|---|---|---|---|
| Cell lysate | ||||
| MKN7 + EBV/MKN7 | 2.147 | 1.051 | ||
| MKN74 + EBV/MKN74 | 4.046 | 2.973 | ||
| Exosome | ||||
| MKN7 + EBV/MKN7 | 0.505/0.515 | not applicable | ||
| 0.981 | 2.104 | 1.601 | ||
| MKN74 + EBV/MKN74 | 0.310/0.380 | 33.9 nm/71.3 nm | ||
| 0.8158 | 2.829 | 2.878 | 0.475 |
| PROTEIN (µG/105 Cell) Fold | Cd63 Density Fold | CD81 Density Fold | Size | |
|---|---|---|---|---|
| Cell lysate | ||||
| SNU719/MKN7 | 2.617 | 1.032 | ||
| SNU719/MKN74 | 2.005 | 3.324 | ||
| Exosome | ||||
| SNU719/MKN7 | 2.29/0.515 | not applicable | ||
| 4.447 | 4.844 | 2.504 | ||
| SNU719/MKN74 | 2.29/0.380 | 69.6 nm/71.3 nm | ||
| 6.026 | 8.785 | 5.605 | 0.976 |
| Variables | EBV-Negative GC | EBVaGC |
|---|---|---|
| N = 40 | N = 41 | |
| Age (y) | ||
| Median (range) | 65 (33–82) | 69 (41–87) |
| Sex | ||
| Female | 7 (17.5) | 6 (14.6%) |
| Male | 33 (82.5%) | 35 (85.4%) |
| Size (cm) | ||
| Median (range) | 5.1 (0.3–16.0) | 4.5 (1.3–12.5) |
| Location | ||
| Upper | 5 (12.5%) | 22 (53.7%) |
| Middle | 21 (52.5%) | 17 (41.5%) |
| Lower | 14 (35.0%) | 2 (4.9%) |
| Lauren classification | ||
| Diffuse | 18 (45.0%) | 20 (48.8%) |
| Intestinal | 22 (55.0%) | 21 (51.2%) |
| Tumor depth | ||
| Early | 21 (52.5%) | 24 (58.5%) |
| Advanced | 19 (47.5%) | 17 (41.5%) |
| EBV-Negative GC | EBVaGC | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Langerin | CD1a | S100 | CD83 | CD86 | BDCA-2 | Langerin | CD1a | S100 | CD83 | CD86 | BDCA-2 | ||
| Age | <65 | 2.71 | 1.30 | 1.21 | 0.13 | 1.32 | 2.88 | 9.98 | 5.40 | 5.96 | 1.58 | 3.01 | 6.49 |
| ≥65 | 1.16 | 1.09 | 1.69 | 0.27 | 0.49 | 2.64 | 7.10 | 4.56 | 3.29 | 0.90 | 3.29 | 7.71 | |
| Sex | Male | 1.75 | 1.29 | 1.64 | 0.23 | 1.01 | 2.84 | 8.42 | 4.84 | 4.18 | 1.16 | 2.51 * | 6.25 |
| Female | 2.61 | 0.69 | 0.61 | 0.07 | 0.50 | 2.37 | 6.58 | 5.07 | 4.79 | 1.10 | 6.65 * | 13.19 | |
| Size | <45 mm | 2.92 | 0.71 | 1.80 | 0.24 | 0.57 | 3.94 | 10.60 | 6.17 | 5.34 | 1.36 | 3.58 | 7.82 |
| ≥45 mm | 1.22 | 1.50 | 1.24 | 0.18 | 1.16 | 1.96 | 5.81 | 3.63 | 3.24 | 0.95 | 2.84 | 6.74 | |
| Lauren classification | Intestinal | 2.22 | 0.75 | 1.81 | 0.19 | 0.63 | 2.52 | 7.00 | 5.42 | 3.63 | 1.05 | 3.61 | 4.73 * |
| Diffuse | 1.51 | 1.72 | 1.04 | 0.21 | 1.25 | 3.04 | 9.36 | 4.30 | 4.93 | 1.25 | 2.78 | 9.93 * | |
| Tumor depth | Early | 1.92 | 0.50 * | 1.92 | 0.18 | 0.49 | 3.05 | 9.35 | 5.12 | 4.31 | 1.56 * | 3.37 | 5.45 * |
| Advanced | 1.88 | 1.94 * | 0.96 | 0.22 | 1.37 | 2.43 | 6.46 | 4.51 | 4.20 | 0.58 * | 2.90 | 9.82 * | |
| Venous invasion | Absent | 2.08 | 0.54 * | 1.39 | 0.30 | 1.15 | 3.81 | 7.87 | 4.91 | 4.04 | 1.27 | 4.14 | 8.39 |
| Present | 1.68 | 1.97 * | 1.56 | 0.09 | 0.64 | 1.46 | 8.35 | 4.84 | 4.43 | 1.07 | 2.53 | 6.47 | |
| Lymphatic invasion | Absent | 2.02 | 0.45 * | 1.83 | 0.18 | 0.51 | 1.52 | 9.58 | 5.84 * | 5.03 | 1.45 | 4.03 | 7.70 |
| Present | 1.78 | 1.93 * | 1.09 | 0.22 | 1.34 | 3.99 | 5.40 | 2.99 * | 2.79 | 0.57 | 1.79 | 6.42 | |
| Lymph node metastasis | Absent | 1.40 * | 0.44 * | 2.03 | 0.21 | 0.50 | 2.98 | 9.04 | 5.44 | 4.78 | 1.35 | 3.72 | 7.12 |
| Present | 2.35 * | 1.86 * | 0.95 | 0.19 | 1.26 | 2.55 | 5.74 | 3.32 | 2.86 | 0.61 | 1.90 | 7.66 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hinata, M.; Kunita, A.; Abe, H.; Morishita, Y.; Sakuma, K.; Yamashita, H.; Seto, Y.; Ushiku, T.; Fukayama, M. Exosomes of Epstein-Barr Virus-Associated Gastric Carcinoma Suppress Dendritic Cell Maturation. Microorganisms 2020, 8, 1776. https://doi.org/10.3390/microorganisms8111776
Hinata M, Kunita A, Abe H, Morishita Y, Sakuma K, Yamashita H, Seto Y, Ushiku T, Fukayama M. Exosomes of Epstein-Barr Virus-Associated Gastric Carcinoma Suppress Dendritic Cell Maturation. Microorganisms. 2020; 8(11):1776. https://doi.org/10.3390/microorganisms8111776
Chicago/Turabian StyleHinata, Munetoshi, Akiko Kunita, Hiroyuki Abe, Yasuyuki Morishita, Kei Sakuma, Hiroharu Yamashita, Yasuyuki Seto, Tetsuo Ushiku, and Masashi Fukayama. 2020. "Exosomes of Epstein-Barr Virus-Associated Gastric Carcinoma Suppress Dendritic Cell Maturation" Microorganisms 8, no. 11: 1776. https://doi.org/10.3390/microorganisms8111776