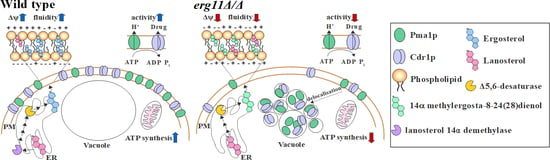
A Crucial Role for Ergosterol in Plasma Membrane Composition, Localisation, and Activity of Cdr1p and H+-ATPase in Candida albicans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Strains and Growth Conditions
2.3. Plasmids and Strains Construction
2.4. Phenotypic Tests and MIC50 Determination
2.5. Real Time PCR
2.6. Isolation of Plasma Membranes
2.7. Sterol Analysis
2.8. Membrane Fluidity Assessment
2.9. Microscopic Studies
2.10. Proton Extrusion Assay
2.11. Di-4-ANEPPS Assay
2.12. ATP Measurements
2.13. Western Blotting
2.14. R6G Efflux Assay
2.15. Statistical Analysis
3. Results
3.1. C. albicans Strain erg11Δ/Δ Is Auxotrophic, Resistant to Azoles and Amphotericin B, and Viable under Aerobic Conditions
3.2. The C. albicans Strain erg11Δ/Δ Accumulates Lanosterol at a Constant Level during Growth, but 14α Methylergosta-8-24(28)dienol at Increasing Level due to High Expression of ERG3 Gene
3.3. In C. albicans Strain erg11Δ/Δ, ATP Concentration, H+-ATPase Activity and Plasma Membrane Potential Are Reduced, and Pma1p Delocalised Earlier to Vacuoles
3.4. In C. albicans Strain erg11Δ/Δ, Cdr1p Is Still Synthesised during Growth but Has Reduced Activity and Is Rapidly Delocalised to the Vacuole
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Odds, F.C. Candida and Candidosis; Saunders, W.B., Ed.; Bailliere Tindall: London, UK, 1988. [Google Scholar]
- Dadar, M.; Tiwari, R.; Karthik, K.; Chakraborty, S.; Shahali, Y.; Dhama, K. Candida albicans—Biology, molecular characterization, pathogenicity, and advances in diagnosis and control—An update. Microb. Pathog. 2018, 117, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.; Lewis, M. Pathogenesis and treatment of oral candidosis. J. Oral Microbiol. 2011, 1, 5771. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.Z.; Yan, L.; Jiang, Y.Y. The synthesis, regulation, and functions of sterols in Candida albicans: Well-known but still lots to learn. Virulence 2016, 7, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, F.; Khodavandi, A.; Zalakian, S. Quantitation of ergosterol content and gene expression profile of ERG11 gene in fluconazole-resistant Candida albicans. Curr. Med. Mycol. 2017, 3, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Gao, N.; Li, C.; Gao, J.; Ying, C. A newly identified amino acid substitution T123I in the 14α-demethylase (Erg11p) of Candida albicans confers azole resistance. FEMS Yeast Res. 2017, 17, fox012. [Google Scholar] [CrossRef]
- Cannon, R.D.; Lamping, E.; Holmes, A.R.; Niimi, K.; Baret, P.V.; Keniya, M.V.; Tanabe, K.; Niimi, M.; Goffeau, A.; Monk, B.C. Efflux-mediated antifungal drug resistance. Clin. Microbiol. Rev. 2009, 22, 291–321. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wu, M.; Wang, Y.; Chen, Y.; Gao, J.; Ying, C. ERG11 couples oxidative stress adaptation, hyphal elongation and virulence in Candida albicans. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef]
- Fonzi, W.A.; Irwin, M.Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics 1993, 134, 717–728. [Google Scholar]
- Szczepaniak, J.; Łukaszewicz, M.; Krasowska, A. Estimation of Candida albicans ABC Transporter Behavior in Real-Time via Fluorescence. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Douglas, L.M.; Wang, H.X.; Keppler-Ross, S.; Dean, N.; Konopka, J.B. Sur7 promotes plasma membrane organization and is needed for resistance to stressful conditions and to the invasive growth and virulence of Candida albicans. mBio 2012, 3. [Google Scholar] [CrossRef]
- Sasse, C.; Schillig, R.; Dierolf, F.; Weyler, M.; Schneider, S.; Mogavero, S.; Rogers, P.D.; Morschhäuser, J. The Transcription Factor Ndt80 Does Not Contribute to Mrr1-, Tac1-, and Upc2-Mediated Fluconazole Resistance in Candida albicans. PLoS ONE 2011, 6, e25623. [Google Scholar] [CrossRef] [PubMed]
- Suchodolski, J.; Feder-Kubis, J.; Krasowska, A. Antifungal activity of ionic liquids based on (−)-menthol: A mechanism study. Microbiol. Res. 2017, 197, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Krasowska, A.; Chmielewska, L.; Prescha, A.; Váchová, L.; Sigler, K. Viability and formation of conjugated dienes in plasma membrane lipids of Saccharomyces cerevisiae, Schizosaccharomyces pombe, Rhodotorula glutinis and Candida albicans exposed to hydrophilic, amphiphilic and hydrophobic pro-oxidants. Folia Microbiol. 2002, 47, 145. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; MacKenzie, A.; Girnun, G.; Del Poeta, M. Analysis of sphingolipids, sterols, and phospholipids in human pathogenic Cryptococcus strains. J. Lipid Res. 2017, 58, 2017–2036. [Google Scholar] [CrossRef] [PubMed]
- Ishmayana, S.; Kennedy, U.J.; Learmonth, R.P. Further investigation of relationships between membrane fluidity and ethanol tolerance in Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 2017, 33, 218. [Google Scholar] [CrossRef]
- Vida, T.A.; Emr, S.D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 1995, 128, 779–792. [Google Scholar] [CrossRef] [PubMed]
- Lachowicz, T.M.; Krasowska, A.; Łuczyński, J.; Witek, S. Plasma membrane H + ATPase activity in wilde type and mutants of yeast Saccharomyces cerevisiae treated by some lysosomotropic drugs. Folia Microbiol. 1998, 43, 203–205. [Google Scholar] [CrossRef]
- Suchodolski, J.; Krasowska, A. Plasma Membrane potential of Candida albicans measured by Di-4-ANEPPS fluorescence depends on growth phase and regulatory factors. Microorganisms 2019, 7, 110. [Google Scholar] [CrossRef]
- Krasowska, A.; Łukaszewicz, M.; Bartosiewicz, D.; Sigler, K. Cell ATP level of Saccharomyces cerevisiae sensitively responds to culture growth and drug-inflicted variations in membrane integrity and PDR pump activity. Biochem. Biophys. Res. Commun. 2010, 395, 51–55. [Google Scholar] [CrossRef]
- Szczepaniak, J.; Cieślik, W.; Romanowicz, A.; Musioł, R.; Krasowska, A. Blocking and dislocation of Candida albicans Cdr1p transporter by styrylquinolines. Int. J. Antimicrob. Agents 2017, 50, 171–176. [Google Scholar] [CrossRef]
- Reuß, O.; Vik, Å.; Kolter, R.; Morschhäuser, J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 2004, 341, 119–127. [Google Scholar] [CrossRef]
- Kumar, A.; Zarychanski, R.; Pisipati, A.; Kumar, A.; Kethireddy, S.; Bow, E.J. Fungicidal versus fungistatic therapy of invasive Candida infection in non-neutropenic adults: A meta-analysis. Mycology 2018, 9, 116–128. [Google Scholar] [CrossRef]
- Oliver, B.G.; Song, J.L.; Choiniere, J.H.; White, T.C. cis-Acting Elements within the Candida albicans ERG11 Promoter Mediate the Azole Response through Transcription Factor Upc2p. Eukaryot. Cell 2007, 6, 2231–2239. [Google Scholar] [CrossRef]
- Song, J.L.; Harry, J.B.; Eastman, R.T.; Oliver, B.G.; White, T.C. The Candida albicans Lanosterol 14-α-Demethylase (ERG11) Gene Promoter Is Maximally Induced after Prolonged Growth with Antifungal Drugs. Antimicrob. Agents Chemother. 2004, 48, 1136–1144. [Google Scholar] [CrossRef]
- Martel, C.M.; Parker, J.E.; Bader, O.; Weig, M.; Gross, U.; Warrilow, A.G.S.; Kelly, D.E.; Kelly, S.L. A clinical isolate of Candida albicans with mutations in ERG11 (encoding sterol 14α-demethylase) and ERG5 (encoding C22 desaturase) is cross resistant to azoles and amphotericin B. Antimicrob. Agents Chemother. 2010, 54, 3578–3583. [Google Scholar] [CrossRef]
- Endress, E.; Bayerl, S.; Prechtel, K.; Maier, C.; Merkel, R.; Bayerl, T.M. The effect of cholesterol, lanosterol, and ergosterol on lecithin bilayer mechanical properties at molecular and microscopic dimensions: A solid-state NMR and micropipet study. Langmuir 2002, 18, 3293–3299. [Google Scholar] [CrossRef]
- Daum, G.; Wagner, A.; Czabany, T.; Athenstaedt, K. Dynamics of neutral lipid storage and mobilization in yeast. Biochimie 2007, 89, 243–248. [Google Scholar] [CrossRef]
- Thomas, E.; Roman, E.; Claypool, S.; Manzoor, N.; Pla, J.; Panwara, S.L. Mitochondria Influence CDR1 Efflux Pump Activity, Hog1-Mediated Oxidative Stress Pathway, Iron Homeostasis, and Ergosterol Levels in Candida albicans. Antimicrob. Agents Chemother. 2013, 57, 5580–5599. [Google Scholar] [CrossRef]
- Qi, Y.; Liu, H.; Yu, J.; Chen, X.; Liu, L. Med15B Regulates Acid Stress Response and Tolerance in Candida glabrata by Altering Membrane Lipid Composition. Appl. Environ. Microbiol. 2017, 83, e01128-17. [Google Scholar] [CrossRef]
- Calahorra, M.; Sanchez, N.S.; Pena, A. Characterization of glycolytic metabolism and ion transport of Candida albicans. Yeast 2012, 29, 357–370. [Google Scholar] [CrossRef]
- Moron, L.; Cabrera, E.C. Detection of azole resistance and ERG11 point mutations in Candida albicans isolates from tertiary hospitals in the Philippines. Curr. Res. Environ. Appl. Mycol. 2018, 8, 298–305. [Google Scholar] [CrossRef]
- Hu, C.; Zhou, M.; Wang, W.; Sun, X.; Yarden, O.; Li, S. Abnormal ergosterol biosynthesis activates transcriptional responses to antifungal azoles. Front. Microbiol. 2018, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Sanglard, D.; Coste, A.; Ferrari, S. Antifungal drug resistance mechanisms in fungal pathogens from the perspective of transcriptional gene regulation. FEMS Yeast Res. 2009, 9, 1029–1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bard, M.; Lees, N.D.; Turi, T.; Craft, D.; Cofrin, L.; Barbuch, R.; Koegel, C.; Loper, J.C. Sterol synthesis and viability of erg11 (cytochrome P450 lanosterol demethylase) mutations in Saccharomyces cerevisiae and Candida albicans. Lipids 1993, 28, 963–967. [Google Scholar] [CrossRef] [PubMed]
- Sanglard, D.; Francoise, I.; Parkinson, T.; Falconer, D.; Jacques, B. Candida albicans Mutations in the Ergosterol Biosynthetic Pathway and Resistance to Several Antifungal Agents. Antimicrob. Agents Chemother. 2003, 47, 2404–2412. [Google Scholar] [CrossRef]
- Geber, A.; Hitchcock, C.A.; Swartz, J.E.; Pullen, F.S.; Marsden, K.E.; Kwon-Chung, K.J.; Bennett, J.E. Deletion of the Candida glabrata ERG3 and ERG11 genes: Effect on cell viability, cell growth, sterol compo- sition, and antifungal susceptibility. Antimicrob. Agents Chemother. 1995, 39, 2708–2717. [Google Scholar] [CrossRef]
- Kelly, S.L.; Lamb, D.C.; Corran, A.J.; Baldwin, B.C.; Kelly, D.E. Mode of action and resistance to azole antifungals associated with the formation of 14 α-methylergosta-8,24(28)-dien-3 beta,6 alpha-diol. Biochem. Biophys. Res. Commun. 1995, 207, 910–915. [Google Scholar] [CrossRef]
- Zavrel, M.; Hoot, S.J.; White, T.C. Comparison of Sterol Import under Aerobic and Anaerobic Conditions in Three Fungal Species, Candida albicans, Candida glabrata, and Saccharomyces cerevisiae. Eukaryot. Cell 2013, 12, 725–738. [Google Scholar] [CrossRef]
- Martel, C.M.; Parker, J.E.; Bader, O.; Weig, M.; Gross, U.; Warrilow, A.G.S.; Rolley, N.; Kelly, D.E.; Kelly, S.L. Identification and characterization of four azole-resistant erg3 mutants of Candida albicans. Antimicrob. Agents Chemother. 2010, 54, 4527–4533. [Google Scholar] [CrossRef]
- Watson, P.F.; Rose, M.E.; Ellis, S.W.; England, H.; Kelly, S.L. Defective sterol C5-6 desaturation and azole resistance: A new hypothesis for the mode of action of azole antifungals. Biochem. Biophys. Res. Commun. 1989, 164, 1170–1175. [Google Scholar] [CrossRef]
- Mondal, M.; Mesmin, B.; Mukherjee, S.; Maxfield, F.R. Sterols Are Mainly in the Cytoplasmic Leaflet of the Plasma Membrane and the Endocytic Recycling Compartment in CHO Cells. Mol. Biol. Cell 2009, 20, 581–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Gisbergen, P.A.; Esseling-Ozdoba, A.; Vos, J.W. Microinjecting FM4-64 validates it as a marker of the endocytic pathway in plants. J. Microsc. 2008, 231, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Rosana, Y.; Yasmon, A.; Lestari, D.C. Overexpression and mutation as a genetic mechanism of fluconazole resistance in Candida albicans isolated from human immunodeficiency virus patients in Indonesia. J. Med. Microbiol. 2015, 64, 1046–1052. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.; Balzi, E.; Banerjee, A.; Khandelwal, N.K. All about CDR transporters: Past, present, and future. Yeast 2018, 36, 223–233. [Google Scholar] [CrossRef] [PubMed]





| Strain | Parent | Genotype | Reference |
|---|---|---|---|
| CAF2-1 | ura3Δ::imm434/URA3 | [9] | |
| ASCa1 | ura3Δ::imm434/ura3Δ::imm434 CDR1/CDR1-yEGFP-URA3 | [10] | |
| YHXW11 | ura3Δ::imm434/ura3Δ::imm434 PMA1/PMA1-GFPγ-URA3 | [11] | |
| KS018 | ASCa1 | ura3Δ::imm434/ura3Δ::imm434 CDR1/CDR1-yEGFP-URA3 ERG11/erg11Δ::SAT1-FLIP | This study |
| KS021 | KS018 | ura3Δ::imm434/ura3Δ::imm434 CDR1/CDR1-yEGFP-URA3 ERG11/erg11Δ::FRT | This study |
| KS023 | KS021 | ura3Δ::imm434/ura3Δ::imm434 CDR1/CDR1-yEGFP-URA3 erg11Δ::SAT1-FLIP/erg11Δ::FRT | This study |
| KS014 | CAF2-1 | ura3Δ::imm434/URA3 ERG11/erg11Δ::SAT1-FLIP | This study |
| KS020 | KS014 | ura3Δ::imm434/URA3 ERG11/erg11Δ::FRT | This study |
| KS028 | KS020 | ura3Δ::imm434/URA3 erg11Δ::SAT1-FLIP/erg11Δ::FRT | This study |
| KS043 | YHXW11 | ura3Δ::imm434/ura3Δ::imm434 PMA1/PMA1-GFPγ-URA3 ERG11/erg11Δ::SAT1-FLIP | This study |
| KS044 | KS043 | ura3Δ::imm434/ura3Δ::imm434 PMA1/PMA1-GFPγ-URA3 ERG11/erg11Δ::FRT | This study |
| KS045 | KS044 | ura3Δ::imm434/ura3Δ::imm434 PMA1/PMA1-GFPγ-URA3 erg11Δ::SAT1-FLIP/erg11Δ::FRT | This study |
| Strain | Time of Culture (h) | Ergosterol | Lanosterol | 14α Methylergosta-8-24(28)dienol | GP |
|---|---|---|---|---|---|
| WT | 8 | 39.11 ± 2.3 | 8.48 ± 0.2 | - | -0.41 ± 0.03 |
| 14 | 65.17 ± 1.1 *** | 5.4 ± 0.6 ** | - | -0.32 ± 0.06 *** | |
| 24 | 6.89 ± 0.2 ** | 3 ± 0.2 *** | - | -0.07 ± 0.05 *** | |
| erg11Δ/Δ | 8 | - | 22.5 ± 1.3 | 23.7 ± 2.4 | -0.18 ± 0.08 *** |
| 14 | - | 21.6 ± 2.4 | 46 ± 3.7 ** | -0.17 ± 0.03 ** | |
| 24 | - | 15.6 ± 1.6* | 61.8 ± 4.3** | -0.07 ± 0.008*** |
| Strain | Time of Culture (h) | ATP (nM/1.4 × 105 cells) |
|---|---|---|
| WT | 8 | 29.3 ± 5.4 |
| 14 | 26.6 ± 2 | |
| 24 | 13.5 ± 1.5 ** | |
| erg11Δ/Δ | 8 | 27.7 ± 2 |
| 14 | 22 ± 1.5 * | |
| 24 | 2.8 ± 0.3 ** |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suchodolski, J.; Muraszko, J.; Bernat, P.; Krasowska, A. A Crucial Role for Ergosterol in Plasma Membrane Composition, Localisation, and Activity of Cdr1p and H+-ATPase in Candida albicans. Microorganisms 2019, 7, 378. https://doi.org/10.3390/microorganisms7100378
Suchodolski J, Muraszko J, Bernat P, Krasowska A. A Crucial Role for Ergosterol in Plasma Membrane Composition, Localisation, and Activity of Cdr1p and H+-ATPase in Candida albicans. Microorganisms. 2019; 7(10):378. https://doi.org/10.3390/microorganisms7100378
Chicago/Turabian StyleSuchodolski, Jakub, Jakub Muraszko, Przemysław Bernat, and Anna Krasowska. 2019. "A Crucial Role for Ergosterol in Plasma Membrane Composition, Localisation, and Activity of Cdr1p and H+-ATPase in Candida albicans" Microorganisms 7, no. 10: 378. https://doi.org/10.3390/microorganisms7100378