Thermus thermophilus as a Source of Thermostable Lipolytic Enzymes
Abstract
:1. Lipolytic Enzymes

1.1. Structure and Mechanism

1.2. Applications

2. Lipolytic Enzymes from Thermophilic Microorganisms
Expression and Production
3. Thermus thermophilus (T. thermophilus)
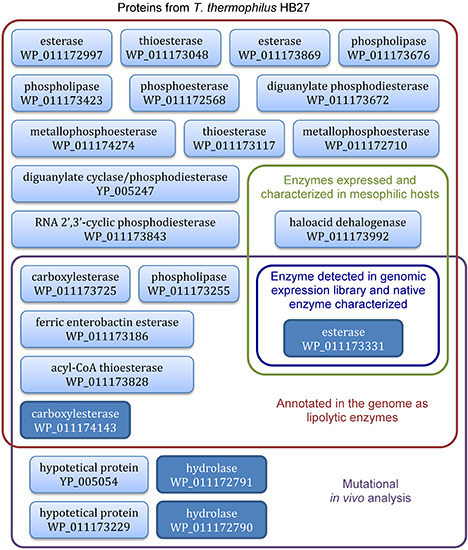
4. Lipolytic Enzymes from Thermus thermophilus HB27
4.1. Heterologous Expression of the 34 kDa T. thermophilus Esterase
| Name | Microorganism | Expression Vector | N-terminus | Optimum pH | Optimum Temperature | Half-life at 85 °C | Extracellular Production (1) | Reference |
|---|---|---|---|---|---|---|---|---|
| E34Tt (Native) | Thermus thermophilus HB27 | - | Full-length | 8.5 | 80 °C | 19 min; 135 min with 1% CHAPS | 42% 120 U/L | [56] |
| ScEST3-O | Saccharomyces cerevisiae BJ3505 | YEpFLAG-1 | ΔN16 | 8.5 | 40 °C | 260 min | <20% 1700 U/L | [59] |
| Km-EP | Kluyveromyces marxianus SLC33 | pSPGK1 | ΔN16 | 8.5–9.6 | 50 °C | 102–108 min | 45% 263 U/L | [58] |
| Km-IN | Kluyveromyces marxianus SLC33 | pNADFL11/INU1 | ΔN16 | 8.5–9.6 | 50 °C | 102–108 min | 50% 328 U/L | [58] |
| Kl-EP | Kluyveromyces lactis PM5-3C | pSPGK1 | ΔN16 | 8.5 | 45 °C | 84–180 min | 12% 345 U/L | [58] |
| Kl-IN | Kluyveromyces lactis PM5-3C | pNADFL11/INU1 | ΔN16 | 8.5 | 45 °C | 84–180 min | 13% 287 U/L | [58] |
| KLEST-3S | Kluyveromyces lactis NRRL-Y1140 | pKLAC1 | ΔN16 | 7.5 | 47.5 °C | 230 min; 5 min with 1% CHAPS | 98% 15000 U/L | [60] |
| KLEST-5A | Kluyveromyces lactis NRRL-Y1140 | pKLAC1 | ΔN26 | 8.5 | 40 °C | 12 min; 2 min with 1% CHAPS | 98% 600 U/L | [60] |
| E34Tt-His6 (2) | Escherichia coli BL21(DE3) | pET-21d(+) | Full-length | 6.3 | 58.2 °C | 107.9 min with 1% CHAPS | <6% <5 U/L | [61] |
4.2. Heterologous Expression of the T. thermophilus Putative Lipase WP_011173992
4.3. Functional Screening of T. thermophilus Lipolytic Enzymes Expressed in E. coli

5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jaeger, K.-E.; Dijkstra, B.W.; Reetz, M.T. Bacterial biocatalysts: Molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu. Rev. Microbiol. 1999, 53, 315–351. [Google Scholar] [CrossRef] [PubMed]
- Bornscheuer, U.T. Microbial carboxyl esterases: Classification, properties and application in biocatalysis. FEMS Microbiol. Rev. 2002, 26, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, S. New tools for exploring old friends-microbial lipases. Appl. Biochem. Biotechnol. 2012, 168, 1163–1196. [Google Scholar] [CrossRef] [PubMed]
- Arpigny, J.L.; Jaeger, K.-E. Bacterial lipolytic enzymes: Classification and properties. Biochem. J. 1999, 343, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-H.; Lee, C.-H.; Oh, T.-K.; Song, J.K.; Yoon, J.-H. Isolation and characterization of a novel lipase from a metagenomic library of tidal flat sediments: Evidence for a new family of bacterial lipases. Appl. Environ. Microbiol. 2006, 72, 7406–7409. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; He, H.; Guo, C.; Sun, B. Identification of two novel esterases from a marine metagenomic library derived from South China Sea. Appl. Microbiol. Biotechnol. 2008, 80, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.-Y.; Oh, K.-H.; Lee, M.-H.; Kang, C.-H.; Oh, T.-K.; Yoon, J.-H. Novel cold-adapted alkaline lipase from an intertidal flat metagenome and proposal for a new family of bacterial lipases. Appl. Environ. Microbiol. 2009, 75, 257–260. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Sun, B. Identification of novel esterase from metagenomic library of Yangtze River. J. Microbiol. Biotechnol. 2009, 19, 187–193. [Google Scholar] [PubMed]
- Fu, C.; Hu, Y.; Xie, F.; Guo, H.; Ashforth, E.J.; Polyak, S.W.; Zhu, B.; Zhang, L. Molecular cloning and characterization of a new cold-active esterase from a deep-sea metagenomic library. Appl. Microbiol. Biotechnol. 2011, 90, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.; Kovacic, F.; Dall Antonia, Y.; Krauss, U.; Fersini, F.; Schmeisser, C.; Lauinger, B.; Bongen, P.; Pietruszka, J.; Schmidt, M.; et al. The metagenome-derived enzymes LipS and LipT increase the diversity of known lipases. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- López-López, O.; Cerdán, M.E.; González-Siso, M.I. Hot spring metagenomics. Life 2013, 3, 308–320. [Google Scholar] [CrossRef] [PubMed]
- López-López, O.; Cerdán, M.E.; González Siso, M.I. New extremophilic lipases and esterases from metagenomics. Curr. Protein Pept. Sci. 2014, 15, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Carr, P.D.; Ollis, D.L. α/β hydrolase fold: An update. Protein Pept. Lett. 2009, 156, 1137–1148. [Google Scholar]
- Turki, S. Towards the development of systems for high-yield production of microbial lipases. Biotechnol. Lett. 2013, 35, 1551–1560. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, A.; Aboul-Enein, H.Y. Application of lipases in kinetic resolution of racemates. Chirality 2005, 17, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Pan, W. Ionic liquids: Green solvents for nonaqueous biocatalysis. Enzyme Microb. Technol. 2005, 37, 19–28. [Google Scholar] [CrossRef]
- Van Rantwijk, F.; Sheldon, R.A. Biocatalysis in ionic liquids. Chem. Rev. 2007, 107, 2757–2785. [Google Scholar] [CrossRef] [PubMed]
- Durand, E.; Lecomte, J.; Villeneuve, P. Deep eutectic solvents: Synthesis, application, and focus on lipase-catalyzed reactions. Eur. J. Lipid Sci. Technol. 2013, 115, 379–385. [Google Scholar] [CrossRef]
- Adlercreutz, P. Immobilisation and application of lipases in organic media. Chem. Soc. Rev. 2013, 42, 6406–6436. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Sharma, B.; Shukla, A.K. Biotechnological approach of microbial lipase: A review. Biotechnology 2011, 10, 23–40. [Google Scholar] [CrossRef]
- Sharma, S.; Kanwar, S.S. Organic Solvent Tolerant Lipases and Applications. Sci. World J. 2014, 2014. [Google Scholar] [CrossRef]
- Salihu, A.; Alam, M.Z.; AbdulKarim, M.I.; Salleh, H.M. Lipase production: An insight in the utilization of renewable agricultural residues. Resour. Conserv. Recycl. 2012, 58, 36–44. [Google Scholar] [CrossRef]
- Haki, G.D.; Rakshit, S.K. Developments in industrially important thermostable enzymes: A review. Bioresour. Technol. 2003, 89, 17–34. [Google Scholar] [CrossRef]
- Andualema, B.; Gessesse, A. Microbial lipases and their industrial applications: Review. Biotechnology 2012, 11, 100–118. [Google Scholar] [CrossRef]
- Bora, L.; Gohain, D.; Das, R. Recent advances in production and biotechnological applications of thermostable and alkaline bacterial lipases. J. Chem. Technol. Biotechnol. 2013, 88, 1959–1970. [Google Scholar] [CrossRef]
- Kumar, S.; Nussinov, R. How do thermophilic proteins deal with heat? Cell. Mol. Life Sci. 2001, 58, 1216–1233. [Google Scholar] [CrossRef] [PubMed]
- Wintrode, P.L.; Zhang, D.; Vaidehi, N.; Arnold, F.H.; Goddard, W.A., III. Protein dynamics in a family of laboratory evolved thermophilic enzymes. J. Mol. Biol. 2003, 327, 745–757. [Google Scholar] [CrossRef]
- Li, W.F.; Zhou, X.X.; Lu, P. Structural features of thermozymes. Biotechnol. Adv. 2005, 23, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Sælensminde, G.; Halskau, Ø., Jr.; Jonassen, I. Amino acid contacts in proteins adapted to different temperatures: Hydrophobic interactions and surface charges play a key role. Extremophiles 2009, 13, 11–20. [Google Scholar]
- Chakravorty, D.; Parameswaran, S.; Dubey, V.K.; Patra, S. In silico characterization of thermostable lipases. Extremophiles 2011, 15, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Elias, M.; Wieczorek, G.; Rosenne, S.; Tawfik, D.S. The universality of enzymatic rate-temperature dependency. Trends Biochem. Sci. 2014, 39, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Cho, A.-R.; Yoo, S.-K.; Kim, E.-J. Cloning, sequencing and expression in Escherichia coli of a thermophilic lipase from Bacillus thermoleovorans ID-1. FEMS Microbiol. Lett. 2000, 186, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Szczesna Antczak, M.; Kubiak, A.; Antczak, T.; Bielecki, S. Enzymatic biodiesel synthesis—Key factors affecting efficiency of the process. Renew. Energy 2009, 34, 1185–1194. [Google Scholar] [CrossRef]
- Schiraldi, C.; de Rosa, M. The production of biocatalysts and biomolecules from extremophiles. Trends Biotechnol. 2002, 20, 515–521. [Google Scholar] [CrossRef]
- Niehaus, F.; Bertoldo, C.; Kähler, M.; Antranikian, G. Extremophiles as a source of novel enzymes for industrial application. Appl. Microbiol. Biotechnol. 1999, 51, 711–729. [Google Scholar] [CrossRef] [PubMed]
- Cava, F.; Hidalgo, A.; Berenguer, J. Thermus thermophilus as biological model. Extremophiles 2009, 13, 213–231. [Google Scholar] [CrossRef] [PubMed]
- Treichel, H.; de Oliveira, D.; Mazutti, M.A.; di Luccio, M.; Oliveira, J.V. A review on microbial lipases production. Food Bioprocess Technol. 2010, 3, 182–196. [Google Scholar] [CrossRef]
- Ming, H.; Yin, Y.-R.; Li, S.; Nie, G.-X.; Yu, T.-T.; Zhou, E.-M.; Liu, L.; Dong, L.; Li, W.-J. Thermus caliditerrae sp. nov., a novel thermophilic species isolated from a geothermal area. Int. J. Syst. Evolut. Microbiol. 2014, 64, 650–656. [Google Scholar]
- Oshima, T.; Imahori, K. Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium from a Japanese thermal spa. Int. J. Syst. Bacteriol. 1974, 24, 102–112. [Google Scholar] [CrossRef]
- Pantazaki, A.A.; Pritsa, A.A.; Kyriakidis, D.A. Biotechnologically relevant enzymes from Thermus thermophilus. Appl. Microbiol. Biotechnol. 2002, 58, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Marteinsson, V.T.; Birrien, J.-L.; Raguénès, G.; Da Costa, M.S.; Prieur, D. Isolation and characterization of Thermus thermophilus Gy1211 from a deep-sea hydrothermal vent. Extremophiles 1999, 3, 247–251. [Google Scholar] [PubMed]
- Henne, A.; Brüggemann, H.; Raasch, C.; Wiezer, A.; Hartsch, T.; Liesegang, H.; Johann, A.; Lienard, T.; Gohl, O.; Martinez-Arias, R.; et al. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 2004, 22, 547–553. [Google Scholar] [PubMed]
- Terpe, K. Overview of thermostable DNA polymerases for classical PCR applications: From molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2013, 97, 10243–10254. [Google Scholar] [CrossRef] [PubMed]
- Rocha-Martín, J.; Vega, D.; Bolivar, J.M.; Godoy, C.A.; Hidalgo, A.; Berenguer, J.; Guisán, J.M.; López-Gallego, F. New biotechnological perspectives of a NADH oxidase variant from Thermus thermophilus HB27 as NAD+-recycling enzyme. BMC Biotechnol. 2011, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeom, S.-J.; Seo, E.-S.; Kim, B.-N.; Kim, Y.-S.; Oh, D.-K. Characterization of a mannose-6-phosphate isomerase from Thermus thermophilus and increased l-ribose production by its R142N mutant. Appl. Environ. Microbiol. 2011, 77, 762–767. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Li, L.; Zheng, Y.; Rui, L.; Hu, C. Perspectives of biotechnological production of l-ribose and its purification. Appl. Microbiol. Biotechnol. 2011, 92, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yin, M.; Zhu, H.; Lu, J.; Cui, Z. Purification and characterization of a hyperthermostable Mn-superoxide dismutase from Thermus thermophilus HB27. Extremophiles 2011, 15, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.-L.; Lee, B.H.; Lacroix, C. Identification of new enzyme activities of several strains of Thermus species. Appl. Microbiol. Biotechnol. 1995, 44, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Domínguez, A.; Sanromán, A.; Fuciños, P.; Rúa, M.L.; Pastrana, L.; Longo, M.A. Quantification of intra- and extra-cellular thermophilic lipase/esterase production by Thermus sp. Biotechnol. Lett. 2004, 26, 705–708. [Google Scholar] [CrossRef] [PubMed]
- Fuciños, P.; Abadín, C.M.; Sanromán, A.; Longo, M.A.; Pastrana, L.; Rúa, M.L. Identification of extracellular lipases/esterases produced by Thermus thermophilus HB27: Partial purification and preliminary biochemical characterisation. J. Biotechnol. 2005, 117, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Fuciños, P.; Pastrana, L.; Sanromán, A.; Longo, M.A.; Hermoso, J.A.; Rúa, M.L. An esterase from Thermus thermophilus HB27 with hyper-thermoalkalophilic properties: Purification, characterisation and structural modelling. J. Mol. Catal. B Enzymatic 2011, 70, 127–137. [Google Scholar] [CrossRef]
- Domínguez, A.; Fuciños, P.; Rúa, M.L.; Pastrana, L.; Longo, M.A.; Sanromán, M.A. Stimulation of novel thermostable extracellular lipolytic enzyme in cultures of Thermus sp. Enzyme Microb. Technol. 2007, 40, 187–194. [Google Scholar] [CrossRef]
- Domínguez, A.; Pastrana, L.; Longo, M.A.; Rúa, M.L.; Sanroman, M.A. Lipolytic enzyme production by Thermus thermophilus HB27 in a stirred tank bioreactor. Biochem. Eng. J. 2005, 26, 95–99. [Google Scholar] [CrossRef]
- Deive, F.J.; Carvalho, E.; Pastrana, L.; Rúa, M.L.; Longo, M.A.; Sanromán, M.A. Assessment of relevant factors influencing lipolytic enzyme production by Thermus thermophilus HB27 in laboratory-scale bioreactors. Chem. Eng. Technol. 2009, 32, 606–612. [Google Scholar] [CrossRef]
- Domínguez, A.; Deive, F.J.; Pastrana, L.; Rúa, M.L.; Longo, M.A.; Sanromán, M.A. Thermostable lipolytic enzymes production in batch and continuous cultures of Thermus thermophilus HB27. Bioprocess Biosyst. Eng. 2010, 33, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Fuciños, P.; Rúa, M.L.; Longo, M.A.; Sanromán, M.A.; Pastrana, L. Thermal spring water enhances lipolytic activity in Thermus thermophilus HB27. Process Biochem. 2008, 43, 1383–1390. [Google Scholar] [CrossRef]
- Deive, F.J.; Carvalho, E.; Pastrana, L.; Rúa, M.L.; Longo, M.A.; Sanroman, M.A. Strategies for improving extracellular lipolytic enzyme production by Thermus thermophilus HB27. Bioresour. Technol. 2009, 100, 3630–3637. [Google Scholar] [CrossRef] [PubMed]
- Rocha, S.N.; Abrahão-Neto, J.; Cerdán, M.E.; Gombert, A.K.; González-Siso, M.I. Heterologous expression of a thermophilic esterase in Kluyveromyces yeasts. Appl. Microbiol. Biotechnol. 2011, 89, 375–385. [Google Scholar] [CrossRef] [PubMed]
- López-López, O.; Fuciños, P.; Pastrana, L.; Rúa, M.L.; Cerdán, M.E.; González-Siso, M.I. Heterologous expression of an esterase from Thermus thermophilus HB27 in Saccharomyces cerevisiae. J. Biotechnol. 2010, 145, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Fuciños, P.; Atanes, E.; López-López, O.; Cerdán, M.E.; González-Siso, M.I.; Pastrana, L.; Luisa Rúa, M. Production and characterization of two N-terminal truncated esterases from Thermus thermophilus HB27 in a mesophilic yeast: Effect of N-terminus in thermal activity and stability. Protein Expr. Purif. 2011, 78, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Fuciños, P.; Atanes, E.; López-López, O.; Solaroli, M.; Cerdán, M.E.; González-Siso, M.I.; Pastrana, L.; Rúa, M.L. Cloning, expression, purification and characterization of an oligomeric His-tagged thermophilic esterase from Thermus thermophilus HB27. Process Biochem. 2014, 49, 927–935. [Google Scholar] [CrossRef]
- López-López, O. Búsqueda de Enzimas Lipolíticas Termófilas y Expresión en Microorganismos Mesófilos; Universidade da Coruña: A Coruña, Spain, 2015. (In Spanish) [Google Scholar]
- Song, C.; Sheng, L.; Zhang, X. Immobilization and Characterization of a Thermostable Lipase. Mar. Biotechnol. 2013, 15, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Du Plessis, E.M.; Berger, E.; Stark, T.; Louw, M.E.; Visser, D. Characterization of a novel thermostable esterase from Thermus scotoductus SA-01: Evidence of a new family of lipolytic esterases. Curr. Microbiol. 2010, 60, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Leis, B.; Angelov, A.; Li, H.; Liebl, W. Genetic analysis of lipolytic activities in Thermus thermophilus HB27. J. Biotechnol. 2014, 191, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Jenney, F.E., Jr.; Adams, M.W.W. The impact of extremophiles on structural genomics (and vice versa). Extremophiles 2008, 12, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Leis, B.; Angelov, A.; Mientus, M.; Li, H.; Pham, V.T.T.; Lauinger, B.; Bongen, P.; Pietruszka, J.; Gonçalves, L.G.; Santos, H.; et al. Identification of novel esterase-active enzymes from hot environments by use of the host bacterium Thermus thermophilus. Front. Microbiol. 2015, 6, 275. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-López, O.; Cerdán, M.-E.; González-Siso, M.-I. Thermus thermophilus as a Source of Thermostable Lipolytic Enzymes. Microorganisms 2015, 3, 792-808. https://doi.org/10.3390/microorganisms3040792
López-López O, Cerdán M-E, González-Siso M-I. Thermus thermophilus as a Source of Thermostable Lipolytic Enzymes. Microorganisms. 2015; 3(4):792-808. https://doi.org/10.3390/microorganisms3040792
Chicago/Turabian StyleLópez-López, Olalla, María-Esperanza Cerdán, and María-Isabel González-Siso. 2015. "Thermus thermophilus as a Source of Thermostable Lipolytic Enzymes" Microorganisms 3, no. 4: 792-808. https://doi.org/10.3390/microorganisms3040792