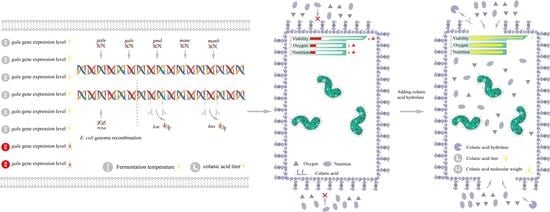
Combinatorial Metabolic Engineering and Enzymatic Catalysis Enable Efficient Production of Colanic Acid
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Plasmids
2.2. Medium and Cultivation
2.3. Expression and Enzyme Activity of Colanic Acid Hydrolase
2.4. Purification and Acid Hydrolysis of Colanic Acid
2.5. Quantification of Colanic Acid
2.6. RNA Extraction and Real-Time PCR
2.7. Enzymatic Hydrolysate Detection
2.8. Atomic Force Microscopy
2.9. Statistical Analysis
3. Results
3.1. Characterization of Colanic Acid
3.2. Relieving the Regulation of the RCS Phosphorylation System on the Synthesis of Colanic Acid and Optimizing the Fermentation Medium
3.3. Enhancement of Colanic Acid Production by Overexpression of Precursors
3.4. Expression and Characterization of Enzymatic Hydrolysates
3.5. Colanic Acid Capsule Layer Inhibits Strain Growth and Glucose Acquisition
3.6. Scale-Up Production of Colanic Acid in a 3-L Bioreactor
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grant, W.D.; Sutherland, I.W.; Wilkinson, J.F. Exopolysaccharide colanic acid and its occurrence in the Enterobacteriaceae. J. Bacteriol. 1969, 100, 1187–1193. [Google Scholar] [CrossRef] [Green Version]
- Ratto, M.; Verhoef, R.; Suihko, M.L.; Blanco, A.; Schols, H.A.; Voragen, A.G.; Wilting, R.; Siika-Aho, M.; Buchert, J. Colanic acid is an exopolysaccharide common to many enterobacteria isolated from paper-machine slimes. J. Ind. Microbiol. Biot. 2006, 33, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Goebel, W.F. Colanic acid. Proc. Natl. Acad. Sci. USA 1963, 49, 464–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaffe, H. The infrared spectral characterization of colanic acid. Proc. Natl. Acad. Sci. USA 1963, 49, 472–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutherland, I.W. Enzymic hydrolysis of colanic acid. Eur. J. Biochem. 1971, 23, 582–587. [Google Scholar] [CrossRef]
- Lawson, C.J.; McCleary, C.W.; Nakada, H.I.; Rees, D.A.; Sutherland, I.W.; Wilkinson, J.F. Structural analysis of colanic acid from Escherichia coli by using methylation and base-catalysed fragmentation. Comparison with polysaccharides from other bacterial sources. Biochem. J. 1969, 115, 947–958. [Google Scholar] [CrossRef] [Green Version]
- Sutherland, I.W. Structural studies on colanic acid, the common exopolysaccharide found in the Enterobacteriaceae, by partial acid hydrolysis. Biochem. J. 1969, 115, 935–945. [Google Scholar] [CrossRef]
- Wu, H.; Chen, S.; Ji, M.; Chen, Q.; Shi, J.; Sun, J. Activation of colanic acid biosynthesis linked to heterologous expression of the polyhydroxybutyrate pathway in Escherichia coli. Int. J. Biol. Macromol. 2019, 128, 752–760. [Google Scholar] [CrossRef]
- Han, B.; Sivaramakrishnan, P.; Lin, C.J.; Neve, I.A.A.; He, J.; Tay, L.W.R.; Sowa, J.N.; Sizovs, A.; Du, G.; Wang, J.; et al. Microbial genetic composition tunes host longevity. Cell 2018, 173, 1058. [Google Scholar] [CrossRef] [Green Version]
- Hartsough, L.A.; Park, M.; Kotlajich, M.V.; Lazar, J.T.; Han, B.; Lin, C.J.; Musteata, E.; Gambill, L.; Wang, M.C.; Tabor, J.J. Optogenetic control of gut bacterial metabolism to promote longevity. Elife 2020, 9, e56849. [Google Scholar] [CrossRef]
- Orskov, I.; Orskov, F.; Jann, B.; Jann, K. Chemistry, and genetics of O and K antigens of Escherichia coli. Bacteriol. Rev. 1977, 41, 667–710. [Google Scholar] [CrossRef]
- Danese, P.N.; Pratt, L.A.; Kolter, R. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 2000, 182, 3593–3596. [Google Scholar] [CrossRef] [Green Version]
- Whitfield, C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 2006, 75, 39–68. [Google Scholar] [CrossRef]
- Majdalani, N.; Gottesman, S. The RCS phosphorelay: A complex signal transduction system. Annu. Rev. Biochem. 2005, 59, 379–405. [Google Scholar] [CrossRef]
- Guo, X.P.; Sun, Y.C. New insights into the non-orthodox two component RCS phosphorelay system. Front. Microbiol. 2017, 8, 2014. [Google Scholar] [CrossRef] [Green Version]
- Rogov, V.V.; Bernhard, F.; Lohr, F.; Dotsch, V. Solution structure of the Escherichia coli YojN histidine-phosphotransferase domain and its interaction with cognate phosphoryl receiver domains. J. Bacteriol. 2004, 343, 1035–1048. [Google Scholar] [CrossRef]
- Brill, J.A.; Quinlan-Walshe, C.; Gottesman, S. Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 1988, 170, 2599–2611. [Google Scholar] [CrossRef] [Green Version]
- Stout, V.; Gottesman, S. RcsB and RcsC: A two-component regulator of capsule synthesis in Escherichia coli. J. Bacteriol. 1990, 172, 659–669. [Google Scholar] [CrossRef] [Green Version]
- Castanie-Cornet, M.P.; Cam, K.; Jacq, A. RcsF is an outer membrane lipoprotein involved in the RcsCDB phosphorelay signaling pathway in Escherichia coli. J. Bacteriol. 2006, 188, 4264–4270. [Google Scholar] [CrossRef] [Green Version]
- Wall, E.A.; Majdalani, N.; Gottesman, S. Igaa negatively regulates the rcs phosphorelay via contact with the RcsD phosphotransfer protein. PLoS Genet. 2020, 16, e1008610. [Google Scholar] [CrossRef]
- Ebel, W.; Trempy, J.E. Escherichia coli RcsA, a positive activator of colanic acid capsular polysaccharide synthesis, functions to activate its own expression. J. Bacteriol. 1999, 181, 577–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.H.; Takeda, S.; Yamada, H.; Ishii, Y.; Yamashino, T.; Mizuno, T. Characterization of the RcsC-->YojN-->RcsB phosphorelay signaling pathway involved in capsular synthesis in Escherichia coli. Biosci. Biotech. Bioch. 2001, 65, 2364–2367. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, C.; Roberts, I.S. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 1999, 31, 1307–1319. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, G.; Andrianopoulos, K.; Hobbs, M.; Reeves, P.R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 1996, 178, 4885–4893. [Google Scholar] [CrossRef] [Green Version]
- Andrianopoulos, K.; Wang, L.; Reeves, P.R. Identification of the fucose synthetase gene in the colanic acid gene cluster of Escherichia coli K-12. J. Bacteriol. 1998, 180, 998–1001. [Google Scholar] [CrossRef] [Green Version]
- Scott, P.M.; Erickson, K.M.; Troutman, J.M. Identification of the functional roles of six key proteins in the biosynthesis of Enterobacteriaceae colanic acid. Biochemistry 2019, 58, 1818–1830. [Google Scholar] [CrossRef]
- Yu, M.; Xu, Y.; Xu, T.; Wang, B.; Sheng, A.; Zhang, X.H. WcaJ, the initiating enzyme for colanic acid synthesis, is required for lipopolysaccharide production, biofilm formation and virulence in Edwardsiella tarda. Aquaculture 2015, 437, 287–291. [Google Scholar] [CrossRef]
- Islam, S.T.; Lam, J.S. Wzx flippase-mediated membrane translocation of sugar polymer precursors in bacteria. Environ. Microbiol. 2013, 15, 1001–1015. [Google Scholar] [CrossRef]
- Reid, A.N.; Whitfield, C. Functional analysis of conserved gene products involved in assembly of Escherichia coli capsules and exopolysaccharides: Evidence for molecular recognition between Wza and Wzc for colanic acid biosynthesis. J. Bacteriol. 2005, 187, 5470–5481. [Google Scholar] [CrossRef] [Green Version]
- Torres-Cabassa, A.S.; Gottesman, S. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J. Bacteriol. 1987, 169, 981–989. [Google Scholar] [CrossRef] [Green Version]
- Sledjeski, D.; Gottesman, S. A small RNA acts as an antisilencer of the H-NS-silenced rcsa gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 1995, 92, 2003–2007. [Google Scholar] [CrossRef] [Green Version]
- Ren, G.; Wang, Z.; Li, Y.; Hu, X.; Wang, X. Effects of lipopolysaccharide core sugar deficiency on colanic acid biosynthesis in Escherichia coli. J. Bacteriol. 2016, 198, 1576–1584. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Zhang, H.; Wang, J.; Chen, S.; Wang, Z.; Zhao, L.; Wang, X. Colanic acid biosynthesis in Escherichia coli is dependent on lipopolysaccharide structure and glucose availability. Microbiol. Res. 2020, 243, 126656. [Google Scholar] [CrossRef]
- Han, H.M.; Kim, I.J.; Yun, E.J.; Lee, J.W.; Cho, Y.; Jin, Y.S.; Kim, K.H. Overproduction of exopolysaccharide colanic acid by Escherichia coli by strain engineering and media optimization. Appl. Biochem. Biotech. 2020, 193, 111–127. [Google Scholar] [CrossRef]
- Sumner, J.B.; Graham, V.A. Dinitrosalicylic acid: A reagent for the estimation of sugar in normal and diabetic urine. J. Biol. Chem. 1921, 47, 5–9. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wu, J.; Deng, X.; Zhang, Y.; Wang, L.; Tian, B.; Xie, B. Application of atomic force microscopy in the study of polysaccharide. J. Ag. Sci. Chn. 2009, 8, 1458–1465. [Google Scholar] [CrossRef]
- Firozi, P.; Zhang, W.; Chen, L.; Quiocho, F.A.; Worley, K.C.; Templeton, N.S. Identification and removal of colanic acid from plasmid DNA preparations: Implications for gene therapy. Gene Ther. 2010, 17, 1484–1499. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Hu, L.; Huang, H.; Wang, H.; Zhang, T.; Chen, J.; Du, G.; Kang, Z. Eliminating the capsule-like layer to promote glucose uptake for hyaluronan production by engineered Corynebacterium glutamicum. Nat. Commun. 2020, 11, 3120. [Google Scholar] [CrossRef]
- Clarke, D.J. The RCS phosphorelay: More than just a two-component pathway. Future Microbiol. 2010, 5, 1173–1184. [Google Scholar] [CrossRef]
- Pristovsek, P.; Sengupta, K.; Lohr, F.; Schafer, B.; von Trebra, M.W.; Ruterjans, H.; Bernhard, F. Structural analysis of the DNA-binding domain of the Erwinia amylovora RcsB protein and its interaction with the RcsAB box. J. Biol. Chem. 2003, 278, 17752–17759. [Google Scholar] [CrossRef] [Green Version]







| Strains | Ratio |
|---|---|
| SR-1 | 2.16 |
| SR-2 | 1.32 |
| SR-3 | 1.14 |
| SR-4 | 2.38 |
| SR-5 | 1.39 |
| SR-6 | 2.41 |
| SR-7 | 3.61 |
| Temperature (°C) | The Average Molecular Weight of Colanic Acid (MDa) |
|---|---|
| 20 | 9.87 |
| 25 | 9.85 |
| 30 | 8.71 |
| 37 | 9.33 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Xu, X.; Lv, X.; Liu, Y.; Li, J.; Du, G.; Liu, L. Combinatorial Metabolic Engineering and Enzymatic Catalysis Enable Efficient Production of Colanic Acid. Microorganisms 2022, 10, 877. https://doi.org/10.3390/microorganisms10050877
Li S, Xu X, Lv X, Liu Y, Li J, Du G, Liu L. Combinatorial Metabolic Engineering and Enzymatic Catalysis Enable Efficient Production of Colanic Acid. Microorganisms. 2022; 10(5):877. https://doi.org/10.3390/microorganisms10050877
Chicago/Turabian StyleLi, Suwei, Xianhao Xu, Xueqin Lv, Yanfeng Liu, Jianghua Li, Guocheng Du, and Long Liu. 2022. "Combinatorial Metabolic Engineering and Enzymatic Catalysis Enable Efficient Production of Colanic Acid" Microorganisms 10, no. 5: 877. https://doi.org/10.3390/microorganisms10050877