Cupriavidus metallidurans CH34 Possesses Aromatic Catabolic Versatility and Degrades Benzene in the Presence of Mercury and Cadmium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Bacterial Strains and Culture Conditions
2.3. Growth of C. metallidurans CH34 on Benzene and Other Aromatic Compounds
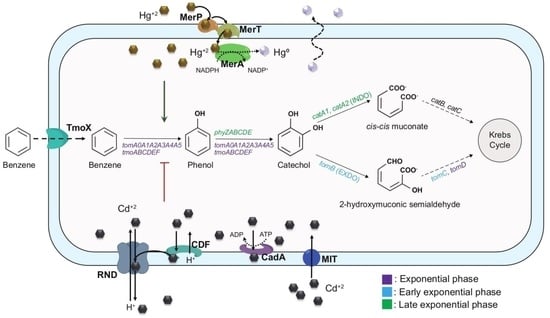
2.4. Aromatic Catabolism Reconstruction of C. metallidurans Strain CH34
2.5. Comparative and Phylogenetic Analysis of Bacterial Multicomponent Monooxygenase (BMM) Gene Clusters
2.6. RNA Isolation and Gene Expression Analysis
2.7. Determination of Mercury and Cadmium Minimum Inhibitory Concentrations
2.8. Benzene Degradation Assays
3. Results
3.1. Genome-Based Reconstruction of the Aromatic Catabolism in Strain CH34
3.2. C. metallidurans CH34 Growth on Aromatic Compounds
3.3. Benzene Catabolism by C. metallidurans CH34
3.4. Transcriptional Analysis during Benzene Degradation
3.5. C. metallidurans CH34 and P. putida F1 growth on Benzene in the Presence of Mercury and Cadmium
3.6. The Effects of Mercury or Cadmium on Benzene Degradation by C. metallidurans CH34 and P. putida F1
4. Discussion
4.1. Aromatic Catabolic Reconstruction of C. metallidurans CH34
4.2. BMMs in C. metallidurans CH34
4.3. Functionality of CH34 BMMs Associated with the Degradation of Benzene
4.4. Effect of Benzene on Bacterial Cadmium MICs
4.5. Effects of Mercury and Cadmium on Benzene Degradation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fuchs, G.; Boll, M.; Heider, J. Microbial degradation of aromatic compounds- From one strategy to four. Nat. Rev. Microbiol. 2011, 9, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, S.; Méndez, V.; Aguila, P.; Seeger, M. Bioremediation of petroleum hydrocarbons: Catabolic genes, microbial communities, and applications. App. Microbiol. Biotech. 2014, 98, 4781–4794. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.H.; Lai, C.Y.; Ye, J.W.; Lin, C.W. Increasing removal of benzene from groundwater using stacked tubular air-cathode microbial fuel cells. J. Clean. Prod. 2018, 194, 78–84. [Google Scholar] [CrossRef]
- van der Perk, M. Soil and Water Contamination, 2nd ed.; Organic Pollutants; CRC Press: Leiden, The Netherland, 2014; p. 167. [Google Scholar]
- Nojiri, H.; Tsuda, M.; Kamagata, Y.; Fukuda, M. Biodegradative Bacteria: How Bacteria Degrade, Survive, Adapt, and Evolve; Springer: Tokyo, Japan, 2014; p. 228. [Google Scholar] [CrossRef]
- Sandrin, T.R.; Maier, R.M.M. Impact of metals on the biodegradation of organic pollutants. Health Perspect. 2003, 111, 1093–1101. [Google Scholar] [CrossRef]
- Sprocati, A.R.; Alisi, C.; Tasso, F.; Marconi, P.; Sciullo, A.; Pinto, V.; Chiavarini, S.; Ubaldi, C.; Cremisini, C. Bioremediation of diesel oil in a co-contaminated soil by bioaugmentation with a microbial formula tailored with native strains selected for heavy metals resistance. Process Biochem. 2012, 47, 1649–1655. [Google Scholar] [CrossRef]
- Nies, D. The biological chemistry of the transition metal transportome of: Cupriavidus metallidurans. Metallomics 2016, 8, 481–507. [Google Scholar] [CrossRef] [Green Version]
- Méndez, V.; Fuentes, S.; Morgante, V.; Hernández, M.; González, M.; Moore, E.; Seeger, M. Novel hydrocarbonoclastic metal-tolerant Acinetobacter and Pseudomonas strains from Aconcagua river oil-polluted soil. J. Soil Sci. Plant Nutr. 2017, 17, 1074–1087. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.X.; Hu, X.; Cao, Y.; Pang, W.J.; Huang, J.Y.; Guo, P.; Huang, L. Biodegradation of phenanthrene and heavy metal removal by acid-tolerant Burkholderia fungorum FM-2. Front. Microbiol. 2019, 10, 408. [Google Scholar] [CrossRef] [Green Version]
- Bravo, G.; Vega-Celedón, P.; Gentina, J.C.; Seeger, M. Bioremediation by Cupriavidus metallidurans strain MSR33 of mercury-polluted agricultural soil in a rotary drum bioreactor and its effects on nitrogen cycle microorganisms. Microorganisms 2020, 8, 1952. [Google Scholar] [CrossRef]
- Gran-Scheuch, A.; Ramos-Zuñiga, J.; Fuentes, E.; Bravo, D.; Pérez-Donoso, J.M. Effect of co-contamination by PAHs and heavy metals on bacterial communities of diesel contaminated soils of South Shetland Islands, Antarctica. Microorganisms 2020, 8, 1749. [Google Scholar] [CrossRef]
- Rizzuti, A.M.; Cohen, A.D.; Stack, E.M. Effects of irradiating peats on their ability to extract BTEX and cadmium from contaminated water. J. Environ. Sci. Health A 1996, 31, 1917–1949. [Google Scholar] [CrossRef]
- Baldrian, P.; in der Wiesche, C.; Jiří, G.; Nerud, F.; Zadražil, F. Influence of cadmium and mercury on activities of ligninolytic enzymes and degradation of polycyclic aromatic hydrocarbons by Pleurotus ostreatus in soil. Appl. Environ. Microbiol. 2000, 66, 2471–2478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavamura, V.N.; Esposito, E. Biotechnological strategies applied to the decontamination of soils polluted with heavy metals. Biotechnol. Adv. 2010, 28, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Ekperusi, O.A.; Aigbodion, I.F. Bioremediation of heavy metals and petroleum hydrocarbons in diesel contaminated soil with the earthworm: Eudrilus eugeniae. SpringerPlus 2015, 4, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joseph, P. Mechanisms of cadmium carcinogenesis. Toxicol. Appl. Pharmacol. 2009, 238, 272–279. [Google Scholar] [CrossRef]
- Begg, S.L.; Eijkelkamp, B.A.; Luo, Z.; Couñago, R.M.; Morey, J.R.; Maher, M.J.; Ong, C.L.Y.; McEwan, A.G.; Kobe, B.; O’Mara, M.L.; et al. Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae. Nat. Commun. 2015, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Kovarik, W. Ethyl-leaded gasoline: How a classic occupational disease became an international public health disaster. Int. J. Occup. Environ. Health 2005, 11, 384–397. [Google Scholar] [CrossRef]
- Kristensen, A.K.B.; Thomsen, J.F.; Mikkelsen, S. A review of mercury exposure among artisanal small-scale gold miners in developing countries. Int. Arch. Occup. Environ. Health 2014, 87, 579–590. [Google Scholar] [CrossRef]
- Bravo, G.; Vega-Celedón, P.; Gentina, J.C.; Seeger, M. Effects of mercury II on Cupriavidus metallidurans strain MSR33 during mercury bioremediation under aerobic and anaerobic conditions. Processes 2020, 8, 893. [Google Scholar] [CrossRef]
- Amor, L.; Kennes, C.; Veiga, M.C. Kinetics of inhibition in the biodegradation of monoaromatic hydrocarbons in presence of heavy metals. Bioresour. Technol. 2001, 78, 181–185. [Google Scholar] [CrossRef] [Green Version]
- Harrison, J.J.; Ceri, H.; Turner, R.J. Multimetal resistance and tolerance in microbial biofilms. Nat. Rev. Microbiol. 2007, 5, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Boyd, E.S.; Barkay, T. The mercury resistance operon: From an origin in a geothermal environment to an efficient detoxification machine. Front. Microbiol. 2012, 3, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montero-Silva, F.; Durán, N.; Seeger, M. Synthesis of extracellular gold nanoparticles using Cupriavidus metallidurans CH34 cells. IET Nanobiotechnol. 2018, 12, 40–46. [Google Scholar] [CrossRef]
- Springael, D.; Diels, L.; Hooyberghs, L.; Kreps, S.; Mergeay, M. Construction and characterization of heavy metal-resistant haloaromatic-degrading Alcaligenes eutrophus strains. Appl. Environ. Microbiol. 1993, 59, 334–339. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Pantoja, D.; De la Iglesia, R.; Pieper, D.H.; González, B. Metabolic reconstruction of aromatic compounds degradation from the genome of the amazing pollutant-degrading bacterium Cupriavidus necator JMP134. FEMS Microbiol. Rev. 2008, 32, 736–794. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Pantoja, D.; Donoso, R.; Agulló, L.; Córdova, M.; Seeger, M.; Pieper, D.H.; González, B. Genomic analysis of the potential for aromatic compounds biodegradation in Burkholderiales. Environ. Microbiol. 2012, 14, 1091–1117. [Google Scholar] [CrossRef]
- Fang, L.C.; Chen, Y.F.; Wang, D.S.; Sun, L.N.; Tang, X.Y.; Hua, R.M. Complete genome sequence of a novel chlorpyrifos degrading bacterium, Cupriavidus nantongensis X1. J. Biotechnol. 2016, 227, 1–2. [Google Scholar] [CrossRef]
- Van Houdt, R.; Provoost, A.; Van Assche, A.; Leys, N.; Lievens, B.; Mijnendonckx, K.; Monsieurs, P. Cupriavidus metallidurans strains with different mobilomes and from distinct environments have comparable phenomes. Genes 2018, 9, 507. [Google Scholar] [CrossRef] [Green Version]
- Moriuchi, R.; Dohra, H.; Kanesaki, Y.; Ogawa, N. Complete genome sequence of 3-chlorobenzoate-degrading bacterium Cupriavidus necator NH9 and reclassification of the strains of the genera Cupriavidus and Ralstonia based on phylogenetic and whole-genome sequence analyses. Front. Microbiol. 2019, 10, 133. [Google Scholar] [CrossRef]
- Alvarez-Santullano, N.; Villegas, P.; Mardones, M.S.; Durán, R.E.; Donoso, R.; González, A.; Sanhueza, C.; Navia, R.; Acevedo, F.; Pérez-Pantoja, D.; et al. Genome-wide metabolic reconstruction of the synthesis of polyhydroxyalkanoates from sugars and fatty acids by Burkholderia sensu lato species. Microorganisms 2021, 9, 1290. [Google Scholar] [CrossRef]
- Suenaga, H.; Yamazoe, A.; Hosoyama, A.; Kimura, N.; Hirose, J.; Watanabe, T.; Fujihara, H.; Futagami, T.; Goto, M.; Furukawa, K. Draft genome sequence of the polychlorinated biphenyl-degrading bacterium Cupriavidus basilensis KF708 (NBRC 110671) isolated from biphenyl-contaminated soil. Genome Announc. 2015, 3, e00143-15. [Google Scholar] [CrossRef] [Green Version]
- Seeger, M.; Saavedra, J.M.; Acevedo, F. Recombinant PCB-degrading bacterium, product for bioremediation and method of application. U.S. Patent 7,989,194 B2, 2 August 2011. [Google Scholar]
- Rojas, L.A.; Yáñez, C.; González, M.; Lobos, S.; Smalla, K.; Seeger, M. Characterization of the metabolically modified heavy metal-resistant Cupriavidus metallidurans strain MSR33 generated for mercury bioremediation. PLoS ONE 2011, 6, e0017555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espinoza-Tofalos, A.; Daghio, M.; González, M.; Papacchini, M.; Franzetti, A.; Seeger, M. Toluene degradation by Cupriavidus metallidurans CH34 in nitrate-reducing conditions and in bioelectrochemical systems. FEMS Microbiol. Lett. 2018, 365, fny119. [Google Scholar] [CrossRef]
- Monchy, S.; Benotmane, M.A.; Janssen, P.; Vallaeys, T.; Taghavi, S.; van der Lelie, D.; Mergeay, M. Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximal viable response to heavy metals. J. Bacteriol. 2007, 189, 7417–7425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janssen, P.J.; Van Houd, T.R.; Moors, H.; Monsieurs, P.; Morin, N.; Michaux, A.; Benotmane, R.; Leys, N.; Vallaeys, T.; Lapidus, A.; et al. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS ONE 2010, 5, e10433. [Google Scholar] [CrossRef]
- Millacura, F.A.; Janssen, P.J.; Monsieurs, P.; Janssen, A.; Provoost, A.; Van Houdt, R.; Rojas, L.A. Unintentional genomic changes endow Cupriavidus metallidurans with an augmented heavy-metal resistance. Genes 2018, 9, 551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alviz-Gazitua, P.; Fuentes-Alburquenque, S.; Rojas, L.A.; Turner, R.J.; Guiliani, N.; Seeger, M. The response of Cupriavidus metallidurans CH34 to cadmium involves inhibition of the initiation of biofilm formation, decrease in intracellular c-di-GMP levels, and a novel metal regulated phosphodiesterase. Front. Microbiol. 2019, 10, 1–17. [Google Scholar] [CrossRef]
- Seeger, M.; Rojas, L.; González, M.; Yáñez, C. Recombinant Bacterium Capable of Removing Mercury (II) Species, Cadmium (II) and Copper (II) in Presence of other Heavy Metals from Polluted Sites, Product for the Bioremediation, Process of Obtaining the Product and Method of Bioremediation. U.S. Patent 8,846,376B2, 30 September 2014. [Google Scholar]
- Mergeay, M.; Nies, D.; Schlegel, H.G.; Gerits, J.; Charles, P.; Van Gijsegem, F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 1985, 162, 328–334. [Google Scholar] [CrossRef] [Green Version]
- Rosier, C.; Leys, N.; Henoumont, C.; Mergeay, M.; Wattiez, R. Purification and characterization of the acetone carboxylase of Cupriavidus metallidurans Strain CH34. Appl. Environ. Microbiol. 2012, 78, 4516–4518. [Google Scholar] [CrossRef] [Green Version]
- Van Houdt, R.; Monsieurs, P.; Mijnendonckx, K.; Provoost, A.; Janssen, A.; Mergeay, M.; Leys, N. Variation in genomic islands contribute to genome plasticity in Cupriavidus metallidurans. BMC Genom. 2012, 13, 111. [Google Scholar] [CrossRef] [Green Version]
- Springael, D.; Diels, L.; Mergeay, M. Transfer and expression of PCB-degradative genes into heavy metal resistant Alcaligenes eutrophus strains. Biodegradation 1994, 5, 343–357. [Google Scholar] [CrossRef] [PubMed]
- David, C.; Daro, A.; Szalai, E.; Atarhouch, T.; Mergeay, M. Formation of polymeric pigments in the presence of bacteria and comparison with chemical oxidative coupling—II. Catabolism of tyrosine and hydroxyphenylacetic acid by Alcaligenes eutrophus CH34 and mutants. Eur. Polym. J. 1996, 32, 669–679. [Google Scholar] [CrossRef]
- Van Houdt, R.; Monchy, S.; Leys, N.; Mergeay, M. New mobile genetic elements in Cupriavidus metallidurans CH34, their possible roles and occurrence in other bacteria. Antonie Leeuwenhoek 2009, 96, 205–226. [Google Scholar] [CrossRef]
- Notomista, E.; Lahm, A.; Di Donato, A.; Tramontano, A. Evolution of bacterial and archaeal multicomponent monooxygenases. J. Mol. Evol. 2003, 56, 435–445. [Google Scholar] [CrossRef]
- Reardon, K.F.; Mosteller, D.C.; Bull Rogers, J.D. Biodegradation kinetics of benzene, toluene, and phenol as single and mixed substrates for Pseudomonas putida F1. Biotechnol. Bioeng. 2000, 69, 385–400. [Google Scholar] [CrossRef]
- Durán, R.E.; Méndez, V.; Rodriguez-Castro, L.; Barra-Sanhueza, B.; Salvà-Serra, F.; Moore, E.R.B.; Castro-Nallar, E.; Seeger, M. Genomic and physiological traits of the marine bacterium Alcaligenes aquatilis QD168 isolated from Quintero Bay, Central Chile, reveal a robust adaptive response to environmental stressors. Front. Microbiol. 2019, 10, 528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, L.C.; Shingle, V.; Sze, C.C.; Poh, C.L. Cloning and sequences of the first eight genes of the chromosomally encoded (methyl) phenol degradation pathway from Pseudomonas putida P35X. Gene 1994, 151, 29–36. [Google Scholar] [CrossRef]
- Reardon, K.F.; Mosteller, D.C.; Rogers Bull, J.D.; DuTeau, N.M.; Kim, K.H. Biodegradation kinetics of aromatic hydrocarbon mixtures by pure and mixed bacterial cultures. Environ. Health Persp. 2002, 110, 1005–1101. [Google Scholar] [CrossRef] [Green Version]
- Sandhu, A.; Halverson, L.J.; Beattie, G.A. Bacterial degradation of airborne phenol in the phyllosphere. Environ. Microbiol. 2007, 9, 383–392. [Google Scholar] [CrossRef]
- Parales, R.E.; Parales, J.V.; Pelletier, D.A.; Ditty, J.L. Chapter 1 Diversity of microbial toluene degradation pathways. Adv. Appl. Microbiol. 2008, 64, 1–73. [Google Scholar] [CrossRef]
- Cai, S.; Chen, L.W.; Ai, Y.C.; Qiu, J.G.; Wang, C.H.; Shi, C.; He, J.; Cai, T.M. Degradation of diphenyl ether in Sphingobium phenoxybenzoativorans SC_3 is initiated by a novel ring cleavage dioxygenase. Appl. Environ. Microbiol. 2017, 83, e00104-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arai, H.; Ishii, M. Draft genome sequence of Comamonas testosteroni TA441, a bacterium that has a cryptic phenol degradation gene cluster. Microbiol. Resour. Announc. 2019, 8, e00946-19. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, S.; Wakem, M.; Dijkman, G.; Alsarraj, M.; Nguyen, M. A practical approach to RT728 qPCR-Publishing data that conform to the MIQE guidelines. Methods 2010, 50, 729. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlation. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Sala-Trepat, J.M.; Evans, W.C. The meta cleavage of catechol by Azotobacter species. 4-Oxalocrotonate pathway. Eur. J. Biochem. 1971, 20, 400–413. [Google Scholar] [CrossRef]
- Hearn, E.M.; Patel, D.R.; Van Den Berg, B. Outer-membrane transport of aromatic hydrocarbons as a first step in biodegradation. Proc. Natl. Acad. Sci. USA 2008, 105, 8601–8606. [Google Scholar] [CrossRef] [Green Version]
- Rüegg, I.; Hafner, T.; Bucheli-Witschel, M.; Egli, T. Dynamics of benzene and toluene degradation in Pseudomonas putida F1 in the presence of the alternative substrate succinate. Eng. Life Sci. 2007, 7, 331–342. [Google Scholar] [CrossRef]
- Yu, H.; Kim, B.J.; Rittmann, B.E. The roles of intermediates in biodegradation of benzene, toluene, and p-xylene by Pseudomonas putida F1. Biodegradation 2001, 12, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Chai, L.; Tang, C.; Yang, Z.; Zhang, H.; Chen, R.; Chen, Y.; Zheng, Y. Characterization and genomic analysis of kraft lignin biodegradation by the beta-proteobacterium Cupriavidus basilensis B-8. Biotechnol. Biofuels. 2013, 6, 1–4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriuchi, R.; Dohra, H.; Kanesaki, Y.; Ogawa, N. Transcriptome differences between Cupriavidus necator NH9 grown with 3-chlorobenzoate and that grown with benzoate. Biosci. Biotechnol. Biochem. 2021, 85, 1546–1561. [Google Scholar] [CrossRef]
- Lykidis, A.; Perez-Pantoja, D.; Ledger, T.; Mavromatis, K.; Anderson, I.J.; Ivanova, N.N.; Hooper, S.D.; Lapidus, A.; Lucas, S.; Gonzalez, B.; et al. The complete multipartite genome sequence of Cupriavidus necator JMP134, a versatile pollutant degrader. PLoS ONE 2010, 5, e9729. [Google Scholar] [CrossRef]
- Sauret-Ignazi, G.; Gagnon, J.; Béguin, C.; Barrelle, M.; Markowicz, Y.; Pelmont, J.; Toussaint, A. Characterization of a chromosomally encoded catechol 1,2-dioxygenase (E.C. 1.13.11.1) from Alcaligenes eutrophus CH34. Arch. Microbiol. 1996, 166, 42–50. [Google Scholar] [CrossRef]
- Han, L.; Chen, S.; Zhou, J. Expression and cloning of catA encoding a catechol 1,2-dioxygenase from the 2,4-D-degrading strain Cupriavidus campinensis BJ71. Prep. Biochem. Biotechnol. 2020, 50, 486–493. [Google Scholar] [CrossRef]
- Pérez-Pantoja, D.; Ledger, T.; Pieper, D.H.; González, B. Efficient turnover of chlorocatechols is essential for growth of Ralstonia eutropha JMP134 (pJP4) in 3-chlorobenzoic acid. J. Bacteriol. 2003, 185, 1534–1542. [Google Scholar] [CrossRef] [Green Version]
- Donoso, R.A.; Ruiz, D.; Gárate-Castro, C.; Villegas, P.; González-Pastor, J.E.; de Lorenzo, V.; González, B.; Pérez-Pantoja, D. Identification of a self-sufficient cytochrome P450 monooxygenase from Cupriavidus pinatubonensis JMP134 involved in 2-hydroxyphenylacetic acid catabolism, via homogentisate pathway. Microb. Biotechnol. 2021, 14, 1944–1960. [Google Scholar] [CrossRef]
- Cafaro, V.; Izzo, V.; Scognamiglio, R.; Notomista, E.; Capasso, P.; Casbarra, A.; Pucci, P.; Di Donato, A. Phenol hydroxylase and toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1: Interplay between Two Enzymes. Appl. Environ. Microbiol. 2004, 70, 2211–2219. [Google Scholar] [CrossRef] [Green Version]
- Shields, M.S.; Montgomery, S.O.; Chapman, P.J.; Cuskey, S.M.; Pritchard, P.H. Novel pathway of toluene catabolism in the trichloroethylene-degrading bacterium G4. App. Environ. Microbiol. 1989, 55, 1624–1629. [Google Scholar] [CrossRef] [Green Version]
- Hur, H.G.; Newman, L.M.; Wackett, L.P.; Sadowsky, M.J. Toluene 2-monooxygenase-dependent growth of Burkholderia cepacia G4/PR1 on diethyl ether. Appl. Environ. Microbiol. 1997, 63, 1606–1609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Sullivan, L.A.; Weightman, A.J.; Jones, T.H.; Marchbank, A.M.; Tiedje, J.M.; Mahenthiralingam, E. Identifying the genetic basis of ecologically and biotechnologically useful functions of the bacterium Burkholderia vietnamiensis. Environ. Microbiol. 2007, 9, 1017–1034. [Google Scholar] [CrossRef] [PubMed]
- Mahendra, S.; Alvarez-Cohen, L. Kinetics of 1,4-dioxane biodegradation by monooxygenase-expressing bacteria. Environ. Sci. Technol. 2006, 40, 5435–5442. [Google Scholar] [CrossRef] [PubMed]
- Clay, K.M.C.; Fox, B.G.; Steffan, R.J. Toluene monooxygenase-catalyzed epoxidation of alkenes. Appl. Environ. Microbiol. 2000, 66, 1877–1882. [Google Scholar] [CrossRef] [Green Version]
- Tao, Y.; Fishman, A.; Bentley, W.E.; Wood, T.K. Oxidation of benzene to phenol, catechol, and 1,2,3-trihydroxybenzene by toluene 4-monooxygenase of Pseudomonas mendocina KR1 and toluene 3-monooxygenase of Ralstonia pickettii PKO1. App. Environ. Microbiol. 2004, 70, 3814–3820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamzah, R.Y.; Al-Baharna, B.S. Catechol ring-cleavage in Pseudomonas cepacia: The simultaneous induction of ortho and meta pathways. Appl. Microbiol. Biotechnol. 1994, 41, 250–256. [Google Scholar] [CrossRef]
- Cao, B.; Geng, A.; Loh, K.C. Induction of ortho and meta-cleavage pathways in Pseudomonas in biodegradation of high benzoate concentration: MS identification of catabolic enzymes. Appl. Microbiol. Biotechnol. 2008, 81, 99–107. [Google Scholar] [CrossRef]
- Von Rozycki, T.; Nies, D. Cupriavidus metallidurans: Evolution of a metal-resistant bacterium. Antonie Van Leeuwenhoek 2009, 96, 115–139. [Google Scholar] [CrossRef]
- Scherer, J.; Nies, D. CzcP is a novel efflux system contributing to transition metal resistance in Cupriavidus metallidurans CH34. Mol. Microbiol. 2009, 73, 601–621. [Google Scholar] [CrossRef]
- Wu, X.; Monchy, S.; Taghavi, S.; Zhu, W.; Ramos, J.; van der Lelie, D. Comparative genomics and functional analysis of niche-specific adaptation in Pseudomonas putida. FEMS Microbio. Rev. 2011, 35, 299–323. [Google Scholar] [CrossRef] [Green Version]
- Agulló, L.; Romero-Silva, M.J.; Domenech, M.; Seeger, M. p-Cymene promotes its catabolism through the p-cymene and the p-cumate pathways, activates a stress response and reduces the biofilm formation in Burkholderia xenovorans LB400. PLoS ONE 2017, 12, e0169544. [Google Scholar] [CrossRef]
- Heipieper, H.J.; Martínez, P.M. Toxicity of hydrocarbons to microorganisms. In Cellular Ecophysiology of Microbe: Hydrocarbon and Lipid Interactions; Krell, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 335–344. [Google Scholar] [CrossRef]
- Nies, D.H. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 1999, 51, 730–750. [Google Scholar] [CrossRef] [PubMed]
- Kivisaar, M. The effect of cellular redox status on the evolvability of new catabolic pathways. Mbio 2018, 9, 10–13. [Google Scholar] [CrossRef] [Green Version]
- Horn, J.M.; Brunke, M.; Deckwer, W.D.; Timmis, K.N. Pseudomonas putida strains which constitutively overexpress mercury resistance for biodetoxification of organomercurial pollutants. Appl. Environ. Microb. 1994, 60, 357–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velázquez, F.; De Lorenzo, V.; Valls, M. The m-xylene biodegradation capacity of Pseudomonas putida mt-2 is submitted to adaptation to abiotic stresses: Evidence from expression profiling of xyl genes. Environ. Microbiol. 2006, 8, 591–602. [Google Scholar] [CrossRef]
- Chillappagari, S.; Seubert, A.; Trip, H.; Kuipers, O.P.; Marahiel, M.A.; Miethke, M. Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J. Bacteriol. 2010, 192, 2512–2524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaVoie, S.P.; Summers, A.O. Transcriptional responses of Escherichia coli during recovery from inorganic or organic mercury exposure. BMC Genom. 2018, 19, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Ren, X.; Fan, B.; Huang, Z.; Wang, W.; Zhou, H.; Lou, Z.; Ding, H.; Lyu, J.; Tan, G. Zinc toxicity and iron-sulfur cluster biogenesis in Escherichia coli. Appl. Environ. Microbiol. 2019, 85, 1–36. [Google Scholar] [CrossRef] [Green Version]
- Ferianc, P.; Farewell, A.; Nyström, T. The cadmium-stress stimulon of Escherichia coli K-12. Microbiology 1998, 144, 1045–1050. [Google Scholar] [CrossRef] [Green Version]
- Helbig, K.; Bleuel, C.; Krauss, G.J.; Nies, D.H. Glutathione and transition-metal homeostasis in Escherichia coli. J. Bacteriol. 2008, 190, 5431–5438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, A.; Crowley, D.E. Global gene expression responses to cadmium toxicity in Escherichia coli. J. Bacteriol. 2005, 187, 3259–3266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guzik, U.; Hupert-Kocurek, K.; Sałek, K.; Wojcieszyńska, D. Influence of metal ions on bioremediation activity of protocatechuate 3,4-dioxygenase from Stenotrophomonas maltophilia KB2. World J. Microbiol. Biotechnol. 2012, 29, 267–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hupert-Kocurek, K.; Saczyńska, A.; Piotrowska-Seget, Z. Cadmium increases catechol 2,3-dioxygenase activity in Variovorax sp. 12S, a metal-tolerant and phenol-degrading strain. Antonie Leeuwenhoek 2013, 104, 845–853. [Google Scholar] [CrossRef]
- Lipscomb, J.D. Mechanism of extradiol aromatic ring-cleaving dioxygenases. Curr. Opin. Struct. Biol. 2008, 18, 644–649. [Google Scholar] [CrossRef] [Green Version]
- Jindrová, E.; Chocová, M.; Demnerová, K.; Brenner, V. Bacterial aerobic degradation of benzene, toluene, ethylbenzene and xylene. Folia Microbiol. 2002, 47, 83–93. [Google Scholar] [CrossRef]







| Carbon Source | C. metallidurans CH34 | |
|---|---|---|
| Monoaromatic hydrocarbons | ||
| Benzene | + | |
| Toluene a | + | |
| Ethylbenzene | ± | |
| o-Xylene | ± | |
| m-Xylene | ± | |
| p-Xylene | ± | |
| p-Cymene | + | |
| Polycyclic hydrocarbons | ||
| Phenanthrene | - | |
| Anthracene | - | |
| Benzoates | ||
| Benzoate (BA) | ++ | |
| 3-Chlorobenzoate (3-CBA) | + | |
| 4-Chlorobenzoate (4-CBA) | - | |
| 3,5-Dichlorobenzoate (3,5-CBA) | ± | |
| 2-Hydroxybenzoate (salicylate) | +++ | |
| 3-Hydroxybenzoate (3-HBA) | +++ | |
| 4-Hydroxybenzoate (4-HBA) | +++ | |
| 2-Aminobenzoate (anthranilate) | - | |
| 4-Aminobenzoate (pABA) | ++ | |
| 4-Isopropylbenzoate (p-cumate) | - | |
| Phenylacetates | ||
| Phenylacetate (PA) | + | |
| 3-Hydroxyphenylacetate (3-HPA) | +++ | |
| 4-Hydroxyphenylacetate (4-HPA) | ++ | |
| Cinnamate | ++ | |
| Vanillin | +++ | |
| Vanillate | + b | |
| Aromatic amino acids | ||
| L-Phenylalanine | ++ | |
| L-Tyrosine | + b | |
| Other aromatic compounds | ||
| Benzamide | - | |
| Nitrobenzene | - | |
| m-Toluate | - | |
| Hydroxyquinol (HQ) | - | |
| Gallate | - | |
| Other non-aromatic compounds | ||
| Succinate | + | |
| Mercury (µM) | Cadmium (mM) | |||||
|---|---|---|---|---|---|---|
| Strain | Succinate | Benzene | Fold Change | Succinate | Benzene | Fold Change |
| C. metallidurans CH34 | 12.5 | 2 | 6.25 | 4 | 0.8 | 5 |
| P. putida F1 | 3.25 | 1 | 3.25 | 4 | 0.4 | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alviz-Gazitua, P.; Durán, R.E.; Millacura, F.A.; Cárdenas, F.; Rojas, L.A.; Seeger, M. Cupriavidus metallidurans CH34 Possesses Aromatic Catabolic Versatility and Degrades Benzene in the Presence of Mercury and Cadmium. Microorganisms 2022, 10, 484. https://doi.org/10.3390/microorganisms10020484
Alviz-Gazitua P, Durán RE, Millacura FA, Cárdenas F, Rojas LA, Seeger M. Cupriavidus metallidurans CH34 Possesses Aromatic Catabolic Versatility and Degrades Benzene in the Presence of Mercury and Cadmium. Microorganisms. 2022; 10(2):484. https://doi.org/10.3390/microorganisms10020484
Chicago/Turabian StyleAlviz-Gazitua, Pablo, Roberto E. Durán, Felipe A. Millacura, Franco Cárdenas, Luis A. Rojas, and Michael Seeger. 2022. "Cupriavidus metallidurans CH34 Possesses Aromatic Catabolic Versatility and Degrades Benzene in the Presence of Mercury and Cadmium" Microorganisms 10, no. 2: 484. https://doi.org/10.3390/microorganisms10020484