Proteomic Study of Response to Copper, Cadmium, and Chrome Ion Stress in Yarrowia lipolytica Strains Isolated from Andean Mine Tailings in Peru
Abstract
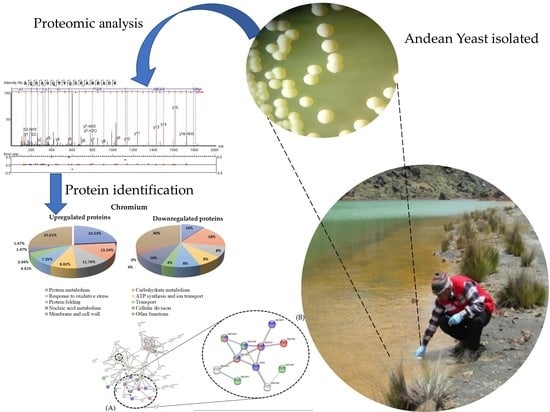
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Sampling and Collection
2.3. Water Physicochemical Analysis and Heavy Metal Measurements
2.4. Yeast Isolation and Culture Conditions
2.5. Heavy Metal Tolerance Determination
2.6. Molecular Identification and Phylogenetic Analysis of Selected Yeasts
2.7. Growth Kinetics and Median Lethal Concentration (LC50) Assay
2.8. Heavy Metal Biosorption
2.9. Protein Extraction
2.10. Digestion and Tagging of Proteins with iTRAQ-8-plex® Reagent
2.11. Mass Spectrometry
2.12. Data Analysis and Database Searching
2.13. Network Analysis
3. Results
3.1. Study Area and Physicochemical Parameters
3.2. Isolation and Molecular Characterization
3.3. Growth and Cytotoxicity Studies against Heavy Metal Ions
3.4. Biosorption Profile
3.5. Effect of Heavy Metal Ions on the Protein Profile
3.6. Identification of Proteins Present in Y. lipolytica under Treatment with Heavy Metal Ions
3.7. Proteins Regulated in Y. lipolytica AMJ6 in the Presence of Cr+6
3.8. Proteins Regulated in Y. lipolytica AMJ6 in the Presence of Cd+2
3.9. Proteins Regulated in Y. lipolytica AMJ6 in the Presence of Cu+2
3.10. Common and Unique Proteins Identified under Treatment with Cr+6, Cd+2, and Cu+2 Ions
3.11. Protein Interaction Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aldenderfer, M.; Craig, N.M.; Speakman, R.J.; Popelka-Filcoff, R. Four-thousand-year-old gold artifacts from the Lake Titicaca basin, southern Peru. Proc. Natl. Acad. Sci. USA 2008, 105, 5002–5005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos-Francés, F.; Martinez-Graña, A.; Alonso Rojo, P.; García Sánchez, A. Geochemical Background and Baseline Values Determination and Spatial Distribution of Heavy Metal Pollution in Soils of the Andes Mountain Range (Cajamarca-Huancavelica, Peru). Int. J. Environ. Res. Public Health 2017, 14, 859. [Google Scholar] [CrossRef] [Green Version]
- Minam, P. Aprueban Estándares de Calidad Ambiental (ECA) para Agua y establecen Disposiciones Complementarias. El Peru 2017, 7, 10–19. [Google Scholar]
- Ferreira da Silva, E.; Zhang, C.; Serrano Pinto, L.s.; Patinha, C.; Reis, P. Hazard assessment on arsenic and lead in soils of Castromil gold mining area, Portugal. Appl. Geochem. 2004, 19, 887–898. [Google Scholar] [CrossRef]
- Fashola, M.O.; Ngole-Jeme, V.M.; Babalola, O.O. Heavy Metal Pollution from Gold Mines: Environmental Effects and Bacterial Strategies for Resistance. Int. J. Environ. Res. Public Health 2016, 13, 1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco-Hernández, M.O.; Vásquez-Murrieta, M.S.; Patiño-Siciliano, A.; Dendooven, L. Heavy metals concentration in plants growing on mine tailings in Central Mexico. Bioresour. Technol. 2010, 101, 3864–3869. [Google Scholar] [CrossRef]
- Mitra, S.; Chakraborty, A.J.; Tareq, A.M.; Emran, T.B.; Nainu, F.; Khusro, A.; Idris, A.M.; Khandaker, M.U.; Osman, H.; Alhumaydhi, F.A.; et al. Impact of heavy metals on the environment and human health: Novel therapeutic insights to counter the toxicity. J. King Saud Univ. Sci. 2022, 34, 101865. [Google Scholar] [CrossRef]
- Briffa, J.; Sinagra, E.; Blundell, R. Heavy metal pollution in the environment and their toxicological effects on humans. Heliyon 2020, 6, e04691. [Google Scholar] [CrossRef]
- Urviola, F.B.S. Determinación de metales pesados en las aguas del río Ananea debido a la actividad minera aurífera, Puno-Perú. Rev. Investig. Esc. Posgrado Una Puno 2009, 5, 5. [Google Scholar]
- Hosiner, D.; Gerber, S.; Lichtenberg-Fraté, H.; Glaser, W.; Schüller, C.; Klipp, E. Impact of acute metal stress in Saccharomyces cerevisiae. PLoS ONE 2014, 9, e83330. [Google Scholar] [CrossRef]
- Llivisaca, S.A.; Burgos, F.A.; Vargas, J.D. Caracterización de Bacterias Metalofijadoras de Mercurío, a Través de la Subunidad 16srna, Mediante la Técnica de pcr-dgge del rio gala (Aguas Abajo en el Recinto San Rafael) en la Parroquia Tenguel. Bachelor’s Thesis, Higher Education Institute Politécnica del Litoral, Guayaquil, Ecuador, 2011. [Google Scholar]
- Orbegozo, J.; Abanto, M.; García, R.; Ramírez, P. Identificación molecular de Pichia guillermondii aislada de aguas ácidas de minas en el Perú y su resistencia a metales pesados. Rev. Peru. Biol. 2008, 15, 91–95. [Google Scholar] [CrossRef] [Green Version]
- Rice, E.W.; Baird, R.B.; Eaton, A.D.; Clesceri, L.S. Standard Methods for the Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 2012; Volume 10. [Google Scholar]
- Protocolo de Monitoreo de la Calidad de los Recursos Hidricos Autoridad Nacional del Agua. 2016. Available online: https://www.ana.gob.pe/sites/default/files/publication/files/protocolo_nacional_para_el_monitoreo_de_la_calidad_de_los_recursos_hidricos_superficiales.pdf (accessed on 10 April 2016).
- Acosta, I.; Moctezuma-Zárate, M.; Gutiérrez, C.; Rodríguez, X. Bioadsorción de Cromo (VI) en Solución Acuosa por la Biomasa Celular de Cryptococcus neoformans y Helminthosporium sp. Inf. Tecnológica 2005, 16, 11–15. [Google Scholar] [CrossRef]
- Irawati, W.; Wijaya, Y.; Christian, S.; Djojo, E.S. Characterization of heavy metals resistant yeast isolated from activated sludge in Rungkut, Surabaya, Indonesia as biosorbent of mercury, copper, and lead. AIP Conf. Proc. 2016, 1744, 020061. [Google Scholar]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic Acids Symposium Series; Information Retrieval Ltd.: London, UK, 1999; pp. 95–98. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Bankar, A.; Zinjarde, S.; Shinde, M.; Gopalghare, G.; Ravikumar, A. Heavy metal tolerance in marine strains of Yarrowia lipolytica. Extremophiles 2018, 22, 617–628. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Wessel, D.; Flügge, U. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984, 138, 141–143. [Google Scholar] [CrossRef]
- Pundir, S.; Martin, M.J.; O’Donovan, C. UniProt protein knowledgebase. In Protein Bioinformatics; Springer: New York, NY, USA, 2017; pp. 41–55. [Google Scholar] [CrossRef] [Green Version]
- Ramos-Fernández, A.; López-Ferrer, D.; Vázquez, J. Improved method for differential expression proteomics using trypsin-catalyzed 18O labeling with a correction for labeling efficiency. Mol. Cell. Proteom. 2007, 6, 1274–1286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mering, C.v.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Hulsen, T.; de Vlieg, J.; Alkema, W. BioVenn—A web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genom. 2008, 9, 488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Chen, C. Biosorption of heavy metals by Saccharomyces cerevisiae: A review. Biotechnol. Adv. 2006, 24, 427–451. [Google Scholar] [CrossRef] [PubMed]
- Sur, I.M.; Micle, V.; Hegyi, A.; Lăzărescu, A.-V. Extraction of Metals from Polluted Soils by Bioleaching in Relation to Environmental Risk Assessment. Materials 2022, 15, 3973. [Google Scholar] [CrossRef]
- Oladipo, O.G.; Awotoye, O.O.; Olayinka, A.; Bezuidenhout, C.C.; Maboeta, M.S. Heavy metal tolerance traits of filamentous fungi isolated from gold and gemstone mining sites. Braz. J. Microbiol. 2018, 49, 29–37. [Google Scholar] [CrossRef]
- Soares, E.V.; Hebbelinck, K.; Soares, H.M. Toxic effects caused by heavy metals in the yeast Saccharomyces cerevisiae: A comparative study. Can. J. Microbiol. 2003, 49, 336–343. [Google Scholar] [CrossRef]
- Codina, J.C.; Pérez-García, A.; Romero, P.; de Vicente, A. A comparison of microbial bioassays for the detection of metal toxicity. Arch. Environ. Contam. Toxicol. 1993, 25, 250–254. [Google Scholar] [CrossRef]
- Wisskirchen, C.; Dold, B.; Friese, K.; Spangenberg, J.E.; Morgenstern, P.; Glaesser, W. Geochemistry of highly acidic mine water following disposal into a natural lake with carbonate bedrock. Appl. Geochem. 2010, 25, 1107–1119. [Google Scholar] [CrossRef]
- Anand, P.; Isar, J.; Saran, S.; Saxena, R.K. Bioaccumulation of copper by Trichoderma viride. Bioresour. Technol. 2006, 97, 1018–1025. [Google Scholar] [CrossRef]
- Bankar, A.; Zinjarde, S.; Telmore, A.; Walke, A.; Ravikumar, A. Morphological response of Yarrowia lipolytica under stress of heavy metals. Can. J. Microbiol. 2018, 64, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.; Bankar, A.; Kumar, A.R.; Gosavi, S.; Zinjarde, S. Removal of hexavalent chromium ions by Yarrowia lipolytica cells modified with phyto-inspired Fe0/Fe3O4 nanoparticles. J. Contam. Hydrol. 2013, 146, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Farhan, S.N.; Khadom, A.A. Biosorption of heavy metals from aqueous solutions by Saccharomyces cerevisiae. Int. J. Ind. Chem. 2015, 6, 119–130. [Google Scholar] [CrossRef] [Green Version]
- Ferdous, A.; Maisha, N.; Sultana, N.; Ahmed, S. Removal of heavy metal from industrial effluents using Baker’s yeast. AIP Conf. Proc. 2016, 1754, 060011. [Google Scholar] [CrossRef]
- Infante, J.C.; De Arco, R.D.; Angulo, M.E. Removal of lead, mercury and nickel using the yeast Saccharomyces cerevisiae. Rev. MVZ Córdoba 2014, 19, 4141–4149. [Google Scholar] [CrossRef] [Green Version]
- Kendrick, B. Fungi: Ecological Importance and Impact on Humans. In eLS; John Wiley & Sons, Ltd.: Chichester, UK, 2011. [Google Scholar] [CrossRef]
- Zieniuk, B.; Fabiszewska, A. Yarrowia lipolytica: A beneficious yeast in biotechnology as a rare opportunistic fungal pathogen: A minireview. World J. Microbiol. Biotechnol. 2018, 35, 10. [Google Scholar] [CrossRef] [Green Version]
- Sumner, E.R.; Shanmuganathan, A.; Sideri, T.C.; Willetts, S.A.; Houghton, J.E.; Avery, S.V. Oxidative protein damage causes chromium toxicity in yeast. Microbiology 2005, 151, 1939–1948. [Google Scholar] [CrossRef] [Green Version]
- Cabiscol, E.; Piulats, E.; Echave, P.; Herrero, E.; Ros, J. Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 27393–27398. [Google Scholar] [CrossRef]
- Muñoz-Esparza, N.C.; Latorre-Moratalla, M.L.; Comas-Basté, O.; Toro-Funes, N.; Veciana-Nogués, M.T.; Vidal-Carou, M.C. Polyamines in Food. Front. Nutr. 2019, 6, 108. [Google Scholar] [CrossRef] [Green Version]
- Chattopadhyay, M.K.; Chen, W.; Poy, G.; Cam, M.; Stiles, D.; Tabor, H. Microarray studies on the genes responsive to the addition of spermidine or spermine to a Saccharomyces cerevisiae spermidine synthase mutant. Yeast 2009, 26, 531–544. [Google Scholar] [CrossRef] [Green Version]
- Hasanuzzaman, M.; Alhaithloul, H.A.S.; Parvin, K.; Bhuyan, M.; Tanveer, M.; Mohsin, S.M.; Nahar, K.; Soliman, M.H.; Mahmud, J.A.; Fujita, M. Polyamine Action under Metal/Metalloid Stress: Regulation of Biosynthesis, Metabolism, and Molecular Interactions. Int. J. Mol. Sci. 2019, 20, 3215. [Google Scholar] [CrossRef] [PubMed]
- Kolhe, N.; Zinjarde, S.; Acharya, C. Impact of uranium exposure on marine yeast, Yarrowia lipolytica: Insights into the yeast strategies to withstand uranium stress. J. Hazard. Mater. 2020, 381, 121226. [Google Scholar] [CrossRef] [PubMed]
- Greetham, D.; Grant, C.M. Antioxidant activity of the yeast mitochondrial one-Cys peroxiredoxin is dependent on thioredoxin reductase and glutathione in vivo. Mol. Cell. Biol. 2009, 29, 3229–3240. [Google Scholar] [CrossRef] [Green Version]
- Mohammadian Fazli, M.; Soleimani, N.; Mehrasbi, M.; Darabian, S.; Mohammadi, J.; Ramazani, A. Highly cadmium tolerant fungi: Their tolerance and removal potential. J. Environ. Health Sci. Eng. 2015, 13, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobson, T.; Priya, S.; Sharma, S.K.; Andersson, S.; Jakobsson, S.; Tanghe, R.; Ashouri, A.; Rauch, S.; Goloubinoff, P.; Christen, P.; et al. Cadmium Causes Misfolding and Aggregation of Cytosolic Proteins in Yeast. Mol. Cell. Biol. 2017, 37, e00490-16. [Google Scholar] [CrossRef] [Green Version]
- Vido, K.; Spector, D.; Lagniel, G.; Lopez, S.; Toledano, M.B.; Labarre, J. A Proteome Analysis of the Cadmium Response in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 8469–8474. [Google Scholar] [CrossRef] [Green Version]
- Le, Q.G.; Ishiwata-Kimata, Y.; Kohno, K.; Kimata, Y. Cadmium impairs protein folding in the endoplasmic reticulum and induces the unfolded protein response. FEMS Yeast Res. 2016, 16, fow049. [Google Scholar] [CrossRef] [Green Version]
- Gardarin, A.; Chédin, S.; Lagniel, G.; Aude, J.C.; Godat, E.; Catty, P.; Labarre, J. Endoplasmic reticulum is a major target of cadmium toxicity in yeast. Mol. Microbiol. 2010, 76, 1034–1048. [Google Scholar] [CrossRef]
- Kirchman, P.A.; Botta, G. Copper supplementation increases yeast life span under conditions requiring respiratory metabolism. Mech. Ageing Dev. 2007, 128, 187–195. [Google Scholar] [CrossRef] [Green Version]
- Rong-Mullins, X.; Winans, M.J.; Lee, J.B.; Lonergan, Z.R.; Pilolli, V.A.; Weatherly, L.M.; Carmenzind, T.W.; Jiang, L.; Cumming, J.R.; Oporto, G.S.; et al. Proteomic and genetic analysis of the response of S. cerevisiae to soluble copper leads to improvement of the antimicrobial function of cellulosic copper nanoparticles. Met. Integr. Biometal Sci. 2017, 9, 1304–1315. [Google Scholar] [CrossRef] [Green Version]
- Sutton, H.C.; Winterbourn, C.C. On the participation of higher oxidation states of iron and copper in fenton reactions. Free. Radic. Biol. Med. 1989, 6, 53–60. [Google Scholar] [CrossRef]
- Miramón, P.; Dunker, C.; Kasper, L.; Jacobsen, I.D.; Barz, D.; Kurzai, O.; Hube, B. A family of glutathione peroxidases contributes to oxidative stress resistance in Candida albicans. Med. Mycol. 2014, 52, 223–239. [Google Scholar] [CrossRef] [PubMed]
- Gross, C.; Kelleher, M.; Iyer, V.R.; Brown, P.O.; Winge, D.R. Identification of the copper regulon in Saccharomyces cerevisiae by DNA microarrays. J. Biol. Chem. 2000, 275, 32310–32316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowman, E.J.; Mandala, S.; Taiz, L.; Bowman, B.J. Structural studies of the vacuolar membrane ATPase from Neurospora crassa and comparison with the tonoplast membrane ATPase from Zea mays. Proc. Natl. Acad. Sci. USA 1986, 83, 48–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eide, D.J.; Bridgham, J.T.; Zhao, Z.; Mattoon, J.R. The vacuolar H(+)-ATPase of Saccharomyces cerevisiae is required for efficient copper detoxification, mitochondrial function, and iron metabolism. Mol. Gen. Genet. 1993, 241, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.Y.; Zhao, Y.; Liu, L.L.; Jia, B.; Zhao, F.; Huang, W.D.; Zhan, J.C. Copper Tolerance and Biosorption of Saccharomyces cerevisiae during Alcoholic Fermentation. PLoS ONE 2015, 10, e0128611. [Google Scholar] [CrossRef]
- Cervantes, C.; Gutierrez-Corona, F. Copper resistance mechanisms in bacteria and fungi. FEMS Microbiol. Rev. 1994, 14, 121–137. [Google Scholar] [CrossRef]
- Kan, G.; Wang, X.; Jiang, J.; Zhang, C.; Chi, M.; Ju, Y.; Shi, C. Copper stress response in yeast Rhodotorula mucilaginosa AN5 isolated from sea ice, Antarctic. MicrobiologyOpen 2019, 8, e00657. [Google Scholar] [CrossRef]
- Lazarova, N.; Krumova, E.; Stefanova, T.; Georgieva, N.; Angelova, M. The oxidative stress response of the filamentous yeast Trichosporon cutaneum R57 to copper, cadmium and chromium exposure. Biotechnol. Biotechnol. Equip. 2014, 28, 855–862. [Google Scholar] [CrossRef]
- Arinbasarova, A.Y.; Biryukova, E.N.; Medentsev, A.G. Antistress systems of the yeast Yarrowia lipolitica (review). Prikl. Biokhimiia I Mikrobiol. 2015, 51, 122–131. [Google Scholar] [CrossRef]
- Ruas, F.A.D.; Barboza, N.R.; Castro-Borges, W.; Guerra-Sá, R. Manganese alters expression of proteins involved in the oxidative stress of Meyerozyma guilliermondii. J. Proteom. 2019, 196, 173–188. [Google Scholar] [CrossRef] [PubMed]















| Sampling Month | Sampling Station | Coordinates UTM 18L | Altitude | pH | Conductivity | TDS 1 | Temperature | |
|---|---|---|---|---|---|---|---|---|
| East | South | (m.a.s.l.) | (µS/cm) | (ppt) | (°C) | |||
| June 2015 | M1 a | 363540.5 | 8814894.33 | 4346 | 2.58 | 4040 | 2.12 | 10.7 |
| M2 a | 363824.88 | 8815541.09 | 4345 | 2.18 | 12,320 | 6.27 | 13.6 | |
| M3 a | 363864.47 | 8815626.03 | 4345 | 3.32 | 9200 | 1.4 | 12.5 | |
| M4 b | 378916.314 | 8772988.85 | 4088 | 7.21 | 320 | 0.16 | 12.7 | |
| M5 b | 374859.529 | 8778313.75 | 4084 | 7.49 | 1050 | 0.52 | 11 | |
| M6 b | 369824.633 | 8781884.24 | 4084 | 7.9 | 430 | 0.21 | 13.5 | |
| M7 b | 363858.889 | 8790283.59 | 4083 | 7.72 | 260 | 0.13 | 13 | |
| November 2015 | M1 a | 363540.5 | 8814894.33 | 4346 | 1.74 | 3999 | 2 | 10 |
| M2 a | 363824.88 | 8815541.09 | 4345 | 1.71 | 3999 | 2 | 10.5 | |
| M3 a | 363864.47 | 8815626.03 | 4345 | 1.73 | 2173 | 1.085 | 11 | |
| M4 b | 378916.314 | 8772988.85 | 4088 | 7.29 | 200 | 0.08 | 8 | |
| M5 b | 374859.529 | 8778313.75 | 4084 | 7.14 | 500 | 0.15 | 11.9 | |
| M6 b | 369824.633 | 8781884.24 | 4084 | 7.24 | 160 | 0.34 | 11 | |
| M7 b | 363858.889 | 8790283.59 | 4083 | 7.33 | 400 | 0.18 | 14 | |
| Metals | Lake Junín—Sampling Station M 5 | * Maximum Permissible Limit 1 (µM) | |
|---|---|---|---|
| June 2015 | November 2015 | ||
| Arsenic | 2.71 | 2.44 | 2 |
| Cadmium | 0.41 | 0.14 | 0.0022 |
| Copper | 66.1 | 11.02 | 1.57 |
| Chromium | 0.21 | 0.21 | 0.21 |
| Nickel | 1.06 | 0.39 | 0.88 |
| Lead | 7.72 | 7.72 | 0.01 |
| Selenium | <0.12 | <0.12 | 0.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Rojas, T.; Espinoza-Culupú, A.; Ramírez, P.; Iwai, L.K.; Montoni, F.; Macedo-Prada, D.; Sulca-López, M.; Durán, Y.; Farfán-López, M.; Herencia, J. Proteomic Study of Response to Copper, Cadmium, and Chrome Ion Stress in Yarrowia lipolytica Strains Isolated from Andean Mine Tailings in Peru. Microorganisms 2022, 10, 2002. https://doi.org/10.3390/microorganisms10102002
Sánchez-Rojas T, Espinoza-Culupú A, Ramírez P, Iwai LK, Montoni F, Macedo-Prada D, Sulca-López M, Durán Y, Farfán-López M, Herencia J. Proteomic Study of Response to Copper, Cadmium, and Chrome Ion Stress in Yarrowia lipolytica Strains Isolated from Andean Mine Tailings in Peru. Microorganisms. 2022; 10(10):2002. https://doi.org/10.3390/microorganisms10102002
Chicago/Turabian StyleSánchez-Rojas, Tito, Abraham Espinoza-Culupú, Pablo Ramírez, Leo Kei Iwai, Fabio Montoni, Diego Macedo-Prada, Marcos Sulca-López, Yerson Durán, Mariella Farfán-López, and Jennifer Herencia. 2022. "Proteomic Study of Response to Copper, Cadmium, and Chrome Ion Stress in Yarrowia lipolytica Strains Isolated from Andean Mine Tailings in Peru" Microorganisms 10, no. 10: 2002. https://doi.org/10.3390/microorganisms10102002