Comparative Degradome Analysis of the Bovine Piroplasmid Pathogens Babesia bovis and Theileria annulata
Abstract
:1. Introduction
2. Results
2.1. Total Degradome Proteinase Repertoire
2.2. Orthologous and Species-Specific Proteinases
2.3. Proteinase Clans and Families
2.4. Proteinases with Two Proteinase Domains
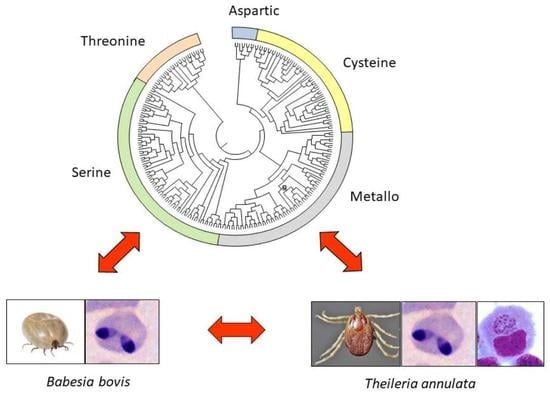
2.5. Molecular Phylogenetic Tree of the Proteinase Domains
2.6. Secreted and Membrane Proteinases
2.7. Ancillary Domains
3. Discussion
4. Materials and Methods
4.1. Mining of the Proteinase Repertoires
4.2. Bioinformatic and Phylogenetic Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, Z. A general review on the prevention and treatment of Theileria annulata in China. Vet. Parasitol. 1997, 70, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Inci, A.; Ica, A.; Yildirim, A.; Vatansever, Z.; Cakmak, A.; Albasan, H.; Cam, Y.; Atasever, A.; Sariozkan, S.; Duzlu, O. Economical impact of tropical theileriosis in the Cappadocia region of Turkey. Parasitol. Res. 2007, 101, S171–S174. [Google Scholar] [CrossRef] [PubMed]
- El Hussein, A.M.; Hassan, S.M.; Salih, D. Current situation of tropical theileriosis in the Sudan. Parasitol. Res. 2012, 111, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Florin-Christensen, M.; Suarez, C.E.; Rodriguez, A.E.; Flores, D.A.; Schnittger, L. Vaccines against bovine babesiosis: Where we are now and possible roads ahead. Parasitology 2014, 141, 1563–1592. [Google Scholar] [CrossRef]
- Florin-Christensen, M.; Schnittger, L.; Bastos, R.G.; Rathinasamy, V.A.; Cooke, B.M.; Alzan, H.F.; Suarez, C.E. Pursuing effective vaccines against cattle diseases caused by apicomplexan protozoa. CAB Rev. 2021, 16, 1–23. [Google Scholar] [CrossRef]
- Pain, A.; Renauld, H.; Berriman, M.; Murphy, L.; Yeats, C.A.; Weir, W.; Kerhornou, A.; Aslett, M.; Bishop, R.; Bouchier, C.; et al. Genome of the Host-Cell Transforming Parasite Theileria annulata Compared with T. Parva. Science 2005, 309, 131–133. [Google Scholar] [CrossRef]
- Brayton, K.A.; Lau, A.O.T.; Herndon, D.R.; Hannick, L.; Kappmeyer, L.S.; Berens, S.J.; Bidwell, S.L.; Brown, W.C.; Crabtree, J.; Fadrosh, D.; et al. Genome Sequence of Babesia bovis and Comparative Analysis of Apicomplexan Hemoprotozoa. PLOS Pathog. 2007, 3, e148. [Google Scholar] [CrossRef]
- Schnittger, L.; Rodriguez, A.E.; Florin-Christensen, M.; Morrison, D.A. Babesia: A world emerging. Infect. Genet. Evol. 2012, 12, 1788–1809. [Google Scholar] [CrossRef]
- Schnittger, L.; Ganzinelli, S.; Bhoora, R.; Omondi, D.; Nijhof, A.M.; Florin-Christensen, M. The Piroplasmida Babesia, Cytauxzoon, and Theileria in farm and companion animals: Species compilation, molecular phylogeny, and evolutionary insights. Parasitol. Res. 2022, 121, 1207–1245. [Google Scholar] [CrossRef]
- Jalovecka, M.; Sojka, D.; Ascencio, M.; Schnittger, L. Babesia Life Cycle—When Phylogeny Meets Biology. Trends Parasitol. 2019, 35, 356–368. [Google Scholar] [CrossRef]
- Florin-Christensen, M.; Schnittger, L. Piroplasmids and ticks: A long-lasting intimate relationship. Front. Biosci. 2009, 35, 3064–3073. [Google Scholar] [CrossRef] [PubMed]
- Ganzinelli, S.; Rodriguez, A.E.; Schnittger, L.; Florin-Christensen, M. Parasitic Protozoa of Farm Animals and Pets; Florin-Christensen, M., Schnittger, L., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 215–239. ISBN 978-3-319-70132-5. [Google Scholar]
- Jalovecka, M.; Hajdusek, O.; Sojka, D.; Kopacek, P.; Malandrin, L. The Complexity of Piroplasms Life Cycles. Front. Cell. Infect. Microbiol. 2018, 8, 248. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.S.; Estrada-Peña, A.; Zintl, A. Vectors of Babesiosis. Annu. Rev. Entomol. 2019, 64, 149–165. [Google Scholar] [CrossRef]
- Hunfeld, K.-P.; Hildebrandt, A.; Gray, J.S. Babesiosis: Recent insights into an ancient disease. Int. J. Parasitol. 2008, 38, 1219–1237. [Google Scholar] [CrossRef] [PubMed]
- Dolan, T.T. Theileriasis: A comprehensive review. Rev. Sci. Tech. Off. Int. Des Epizoot. 1989, 8, 11–78. [Google Scholar] [CrossRef]
- Kiara, H.; Steinaa, L.; Vishvanath, N.; Svitek, N. Babesia in domestic ruminants. In Parasitic Protozoa of Farm Animals and Pets, 1st ed.; Florin-Christensen, M., Schnittger, L., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 187–214. ISBN 978-3-319-70132-5. [Google Scholar]
- Spooner, R.L.; Innes, E.A.; Glass, E.J.; Brown, C.G. Theileria annulata and T. parva infect and transform different bovine mon-onuclear cells. Immunology 1989, 66, 284–288. [Google Scholar] [PubMed]
- Tajeri, S.; Haidar, M.; Sakura, T.; Langsley, G. Interaction between transforming Theileria parasites and their host bovine leukocytes. Mol. Microbiol. 2021, 115, 860–869. [Google Scholar] [CrossRef]
- Sivakumar, T.; Hayashida, K.; Sugimoto, C.; Yokoyama, N. Evolution and genetic diversity of Theileria. Infect. Genet. Evol. 2014, 27, 250–263. [Google Scholar] [CrossRef]
- Shaw, M.K. Cell invasion by Theileria sporozoites. Trends Parasitol. 2003, 19, 2–6. [Google Scholar] [CrossRef]
- Rosenthal, P.; Sijwali, P.; Singh, A.; Shenai, B. Cysteine Proteases of Malaria Parasites: Targets for Chemotherapy. Curr. Pharm. Des. 2002, 8, 1659–1672. [Google Scholar] [CrossRef]
- Yang, G.; Li, J.; Zhang, X.; Zhao, Q.; Liu, Q.; Gong, P. Eimeria tenella: Construction of a recombinant fowlpox virus expressing rhomboid gene and its protective efficacy against homologous infection. Exp. Parasitol. 2008, 119, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Fedeli, C.E.C.; Ferreira, J.H.L.; Mussalem, J.S.; Longo-Maugéri, I.M.; Gentil, L.G.; dos Santos, M.R.M.; Katz, S.; Barbiéri, C.L. Partial protective responses induced by a recombinant cysteine proteinase from Leishmania (Leishmania) amazonensis in a murine model of cutaneous leishmaniasis. Exp. Parasitol. 2010, 124, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Kuang, R.; Gu, J.; Wang, Y. Proteases in Malaria Parasites—A Phylogenomic Perspective. Curr. Genom. 2011, 12, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Pandey, K.C. Centenary celebrations article: Cysteine proteases of human malaria parasites. J. Parasit. Dis. 2011, 35, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zheng, J.; Gong, P.; Zhang, X. Efficacy of Eimeria tenella rhomboid-like protein as a subunit vaccine in protective immunity against homologous challenge. Parasitol. Res. 2012, 110, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- Silva-Almeida, M.; Pereira, B.A.S.; Ribeiro-Guimarães, M.L.; Alves, C.R. Proteinases as virulence factors in Leishmania spp. infection in mammals. Parasit. Vectors 2012, 5, 160. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.-Z.; Xu, Y.; Wang, M.; Petersen, E.; Chen, J.; Huang, S.-Y.; Zhu, X.-Q. Protective efficacy of two novel DNA vaccines expressing Toxoplasma gondii rhomboid 4 and rhomboid 5 proteins against acute and chronic toxoplasmosis in mice. Expert Rev. Vaccines 2015, 14, 1289–1297. [Google Scholar] [CrossRef]
- Rawlings, N.D. Twenty-five years of nomenclature and classification of proteolytic enzymes. Biochim. Biophys. Acta Proteins Proteom. 2019, 1868, 140345. [Google Scholar] [CrossRef]
- Klemba, M.; Goldberg, D.E. Biological Roles of Proteases in Parasitic Protozoa. Annu. Rev. Biochem. 2002, 71, 275–305. [Google Scholar] [CrossRef] [PubMed]
- Lilburn, T.G.; Cai, H.; Zhou, Z.; Wang, Y. Protease-associated cellular networks in malaria parasite Plasmodium falciparum. BMC Genom. 2011, 12 (Suppl. 5), S9. [Google Scholar] [CrossRef] [Green Version]
- Sojka, D.; Šnebergerová, P.; Robbertse, L. Protease Inhibition—An Established Strategy to Combat Infectious Diseases. Int. J. Mol. Sci. 2021, 22, 5762. [Google Scholar] [CrossRef] [PubMed]
- Harris, P.K.; Yeoh, S.; Dluzewski, A.R.; A O’Donnell, R.; Withers-Martinez, C.; Hackett, F.; Bannister, L.H.; Mitchell, G.H.; Blackman, M.J. Molecular Identification of a Malaria Merozoite Surface Sheddase. PLOS Pathog. 2005, 1, e29. [Google Scholar] [CrossRef] [PubMed]
- Yeoh, S.; O’Donnell, R.A.; Koussis, K.; Dluzewski, A.R.; Ansell, K.H.; Osborne, S.A.; Hackett, F.; Withers-Martinez, C.; Mitchell, G.H.; Bannister, L.H.; et al. Subcellular Discharge of a Serine Protease Mediates Release of Invasive Malaria Parasites from Host Erythrocytes. Cell 2007, 131, 1072–1083. [Google Scholar] [CrossRef] [PubMed]
- Koussis, K.; Withers-Martinez, C.; Yeoh, S.; Child, M.; Hackett, F.; Knuepfer, E.; Juliano, L.; Woehlbier, U.; Bujard, H.; Blackman, M.J. A multifunctional serine protease primes the malaria parasite for red blood cell invasion. EMBO J. 2009, 28, 725–735. [Google Scholar] [CrossRef]
- Šnebergerová, P.; Bartošová-Sojková, P.; Jalovecká, M.; Sojka, D. Plasmepsin-like Aspartyl Proteases in Babesia. Pathogens 2021, 10, 1241. [Google Scholar] [CrossRef]
- Sim, B.K.L.; Chitnis, C.E.; Wasniowska, K.; Hadley, T.J.; Miller, L.H. Receptor and Ligand Domains for Invasion of Erythrocytes by Plasmodium falciparum. Science 1994, 264, 1941–1944. [Google Scholar] [CrossRef]
- Baker, R.P.; Wijetilaka, R.; Urban, S. Two Plasmodium Rhomboid Proteases Preferentially Cleave Different Adhesins Implicated in All Invasive Stages of Malaria. PLOS Pathog. 2006, 2, e113. [Google Scholar] [CrossRef]
- O’Donnell, R.A.; Hackett, F.; Howell, S.A.; Treeck, M.; Struck, N.; Krnajski, Z.; Withers-Martinez, C.; Gilberger, T.-W.; Blackman, M.J. Intramembrane proteolysis mediates shedding of a key adhesin during erythrocyte invasion by the malaria parasite. J. Cell Biol. 2006, 174, 1023–1033. [Google Scholar] [CrossRef]
- Urban, S. Making the cut: Central roles of intramembrane proteolysis in pathogenic microorganisms. Nat. Rev. Microbiol. 2009, 7, 411. [Google Scholar] [CrossRef]
- Strisovsky, K. Rhomboid protease inhibitors: Emerging tools and future therapeutics. Semin. Cell Dev. Biol. 2016, 60, 52–62. [Google Scholar] [CrossRef] [Green Version]
- Tichá, A.; Collis, B.; Strisovsky, K. The Rhomboid Superfamily: Structural Mechanisms and Chemical Biology Opportunities. Trends Biochem. Sci. 2018, 43, 726–739. [Google Scholar] [CrossRef]
- Gandhi, S.; Baker, R.P.; Cho, S.; Stanchev, S.; Strisovsky, K.; Urban, S. Designed Parasite-Selective Rhomboid Inhibitors Block Invasion and Clear Blood-Stage Malaria. Cell Chem. Biol. 2020, 27, 1410–1424.e6. [Google Scholar] [CrossRef] [PubMed]
- Francis, S.E.; Sullivan, D.J.; Goldberg, D.E. Hemoglobin metabolism in the malaria parasite Plasmodium falciparum. Annu. Rev. Microbiol. 1997, 51, 97–123. [Google Scholar] [CrossRef]
- Coombs, G.H.; Goldberg, D.E.; Klemba, M.; Berry, C.; Kay, J.; Mottram, J. Aspartic proteases of Plasmodium falciparum and other parasitic protozoa as drug targets. Trends Parasitol. 2001, 17, 532–537. [Google Scholar] [CrossRef]
- Hanspal, M.; Dua, M.; Takakuwa, Y.; Chishti, A.H.; Mizuno, A. Plasmodium falciparum cysteine protease falcipain-2 cleaves erythrocyte membrane skeletal proteins at late stages of parasite development. Blood 2002, 100, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Gluzman, I.Y.; Drew, M.E.; Goldberg, D.E. The Role of Plasmodium falciparum Food Vacuole Plasmepsins. J. Biol. Chem. 2004, 280, 1432–1437. [Google Scholar] [CrossRef] [PubMed]
- Sijwali, P.S.; Rosenthal, P.J. Gene disruption confirms a critical role for the cysteine protease falcipain-2 in hemoglobin hydrolysis by Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 2004, 101, 4384–4389. [Google Scholar] [CrossRef]
- Arastu-Kapur, S.; Ponder, E.L.; Fonović, U.P.; Yeoh, S.; Yuan, F.; Fonovic, M.; Grainger, M.; Phillips, C.I.; Powers, J.C.; Bogyo, M. Identification of proteases that regulate erythrocyte rupture by the malaria parasite Plasmodium falciparum. Nat. Chem. Biol. 2008, 4, 203–213. [Google Scholar] [CrossRef]
- Okubo, K.; Yokoyama, N.; Govind, Y.; Alhassan, A.; Igarashi, I. Babesia bovis: Effects of cysteine protease inhibitors on in vitro growth. Exp. Parasitol. 2007, 117, 214–217. [Google Scholar] [CrossRef]
- Aboge, G.O.; Cao, S.; Terkawi, M.A.; Masatani, T.; Goo, Y.; AbouLaila, M.; Nishikawa, Y.; Igarashi, I.; Suzuki, H.; Xuan, X. Molecular Characterization of Babesia bovis M17 Leucine Aminopeptidase and Inhibition of Babesia Growth by Bestatin. J. Parasitol. 2015, 101, 536–541. [Google Scholar] [CrossRef]
- Munkhjargal, T.; Ishizaki, T.; Guswanto, A.; Takemae, H.; Yokoyama, N.; Igarashi, I. Molecular and biochemical characterization of methionine aminopeptidase of Babesia bovis as a potent drug target. Vet. Parasitol. 2016, 221, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Sojka, D.; Jalovecká, M.; Perner, J. Babesia, Theileria, Plasmodium and Hemoglobin. Microorganisms 2022, 10, 1651. [Google Scholar] [CrossRef] [PubMed]
- Mesplet, M.; Palmer, G.H.; Pedroni, M.J.; Echaide, I.; Florin-Christensen, M.; Schnittger, L.; Lau, A.O. Genome-wide analysis of peptidase content and expression in a virulent and attenuated Babesia bovis strain pair. Mol. Biochem. Parasitol. 2011, 179, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Florin-Christensen, M.; Wieser, S.N.; Suarez, C.E.; Schnittger, L. In Silico Survey and Characterization of Babesia microti Functional and Non-Functional Proteases. Pathogens 2021, 10, 1457. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Bateman, A. Origins of peptidases. Biochimie 2019, 166, 4–18. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Finn, R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2015, 44, D343–D350. [Google Scholar] [CrossRef]
- López-Otín, C.; Bond, J.S. Proteases: Multifunctional Enzymes in Life and Disease. J. Biol. Chem. 2008, 283, 30433–30437. [Google Scholar] [CrossRef]
- E Todd, A.; A Orengo, C.; Thornton, J.M. Sequence and Structural Differences between Enzyme and Nonenzyme Homologs. Structure 2002, 10, 1435–1451. [Google Scholar] [CrossRef]
- Pils, B.; Schultz, J. Inactive Enzyme-homologues Find New Function in Regulatory Processes. J. Mol. Biol. 2004, 340, 399–404. [Google Scholar] [CrossRef]
- Reynolds, S.L.; Fischer, K. Pseudoproteases: Mechanisms and function. Biochem. J. 2015, 468, 17–24. [Google Scholar] [CrossRef]
- Adrain, C.; Cavadas, M. The complex life of rhomboid pseudoproteases. FEBS J. 2020, 287, 4261–4283. [Google Scholar] [CrossRef] [PubMed]
- Mishra, L.; Funk, C. The FtsHi Enzymes of Arabidopsis thaliana: Pseudo-Proteases with an Important Function. Int. J. Mol. Sci. 2021, 22, 5917. [Google Scholar] [CrossRef] [PubMed]
- Zupanič, N.; Počič, J.; Leonardi, A.; Šribar, J.; Kordiš, D.; Križaj, I. Serine pseudoproteases in physiology and disease. FEBS J. 2022. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. Orthologs, Paralogs, and Evolutionary Genomics. Annu. Rev. Genet. 2005, 39, 309–338. [Google Scholar] [CrossRef] [PubMed]
- Gallenti, R.; Poklepovich, T.; Florin-Christensen, M.; Schnittger, L. The repertoire of serine rhomboid proteases of piroplasmids of importance to animal and human health. Int. J. Parasitol. 2021, 51, 455–462. [Google Scholar] [CrossRef]
- Dowse, T.J.; Koussis, K.; Blackman, M.J.; Soldati-Favre, D. Roles of Proteases during Invasion and Egress by Plasmodium and Toxoplasma. Subcell. Biochem. 2008, 47, 121–139. [Google Scholar] [CrossRef]
- Gallenti, R.; Hussein, H.E.; Alzan, H.F.; Suarez, C.E.; Ueti, M.; Asurmendi, S.; Benitez, D.; Araujo, F.R.; Rolls, P.; Sibeko-Matjila, K.; et al. Unraveling the Complexity of the Rhomboid Serine Protease 4 Family of Babesia bovis Using Bioinformatics and Experimental Studies. Pathogens 2022, 11, 344. [Google Scholar] [CrossRef]
- Nejatfard, A.; Wauer, N.; Bhaduri, S.; Conn, A.; Gourkanti, S.; Singh, N.; Kuo, T.; Kandel, R.; Amaro, R.E.; Neal, S.E. Derlin rhomboid pseudoproteases employ substrate engagement and lipid distortion to enable the retrotranslocation of ERAD membrane substrates. Cell Rep. 2021, 37, 109840. [Google Scholar] [CrossRef]
- Sajid, M.; Blackman, M.J.; Doyle, P.; He, C.; Land, K.M.; Lobo, C.; Mackey, Z.; Ndao, M.; Reed, S.L.; Shiels, B.; et al. Antipara-sitic and Antibacterial Drug Discovery: From Molecular Targets to Drug Candidates, 1st ed.; Selzer, P.M., Ed.; Wiley-VCH: Weinheim, Germany, 2009; pp. 190–197. ISBN 978-3-527-32327-2. [Google Scholar]
- Mesplet, M.; Echaide, I.; Dominguez, M.; Mosqueda, J.J.; E Suarez, C.; Schnittger, L.; Florin-Christensen, M. Bovipain-2, the falcipain-2 ortholog, is expressed in intraerythrocytic stages of the tick-transmitted hemoparasite Babesia bovis. Parasit. Vectors 2010, 3, 113. [Google Scholar] [CrossRef]
- Ascencio, M.E.; Florin-Christensen, M.; Mamoun, C.B.; Weir, W.; Shiels, B.; Schnittger, L. Cysteine Proteinase C1A Paralog Profiles Correspond with Phylogenetic Lineages of Pathogenic Piroplasmids. Vet. Sci. 2018, 5, 41. [Google Scholar] [CrossRef] [Green Version]
- Rosenthal, P.J. Cysteine proteases of malaria parasites. Int. J. Parasitol. 2004, 34, 1489–1499. [Google Scholar] [CrossRef] [PubMed]
- Martins, T.M.; Rosário, V.E.D.; Domingos, A. Identification of papain-like cysteine proteases from the bovine piroplasm Babesia bigemina and evolutionary relationship of piroplasms C1 family of cysteine proteases. Exp. Parasitol. 2011, 127, 184–194. [Google Scholar] [CrossRef]
- Carletti, T.; Barreto, C.; Mesplet, M.; Mira, A.; Weir, W.; Shiels, B.; Oliva, A.G.; Schnittger, L.; Florin-Christensen, M. Characterization of a papain-like cysteine protease essential for the survival of Babesia ovis merozoites. Ticks Tick-Borne Dis. 2016, 7, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Pérez, B.; Antunes, S.; Gonçalves, L.M.; Domingos, A.; Gomes, J.R.B.; Gomes, P.; Teixeira, C. Toward the discovery of inhibitors of babesipain-1, a Babesia bigemina cysteine protease: In vitro evaluation, homology modeling and molecular docking studies. J. Comput. Aided Mol. Des. 2013, 27, 823–835. [Google Scholar] [CrossRef] [PubMed]
- Meetei, P.A.; Rathore, R.S.; Prabhu, N.P.; Vindal, V. Modeling of babesipain-1 and identification of natural and synthetic leads for bovine babesiosis drug development. J. Mol. Model. 2016, 22, 1–14. [Google Scholar] [CrossRef]
- Lu, S.; Ascencio, M.E.; Torquato, R.J.; Florin-Christensen, M.; Tanaka, A.S. Kinetic characterization of a novel cysteine peptidase from the protozoan Babesia bovis, a potential target for drug design. Biochimie 2020, 179, 127–134. [Google Scholar] [CrossRef]
- Sonenshine, D.E.; Macaluso, K.R. Microbial Invasion vs. Tick Immune Regulation. Front. Cell. Infect. Microbiol. 2017, 7, 390. [Google Scholar] [CrossRef]
- Zhou, J.; Ueda, M.; Umemiya, R.; Battsetseg, B.; Boldbaatar, D.; Xuan, X.; Fujisaki, K. A secreted cystatin from the tick Haemaphysalis longicornis and its distinct expression patterns in relation to innate immunity. Insect Biochem. Mol. Biol. 2006, 36, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; da Rocha, L.A.; Torquato, R.J.; Jr, I.D.S.V.; Florin-Christensen, M.; Tanaka, A.S. A novel type 1 cystatin involved in the regulation of Rhipicephalus microplus midgut cysteine proteases. Ticks Tick-Borne Dis. 2020, 11, 101374. [Google Scholar] [CrossRef]
- Wei, N.; Du, Y.; Lu, J.; Zhou, Y.; Cao, J.; Zhang, H.; Gong, H.; Zhou, J. A cysteine protease of Babesia microti and its interaction with tick cystatins. Parasitol. Res. 2020, 119, 3013–3022. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- AbouLaila, M.; Nakamura, K.; Govind, Y.; Yokoyama, N.; Igarashi, I. Evaluation of the in vitro growth-inhibitory effect of epoxomicin on Babesia parasites. Vet. Parasitol. 2010, 167, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Jalovecka, M.; Hartmann, D.; Miyamoto, Y.; Eckmann, L.; Hajdusek, O.; O’Donoghue, A.J.; Sojka, D. Validation of Babesia proteasome as a drug target. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Montero, E.; Rafiq, S.; Heck, S.; Lobo, C.A. Inhibition of human erythrocyte invasion by Babesia divergens using serine protease inhibitors. Mol. Biochem. Parasitol. 2007, 153, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Spaccapelo, R.; Janse, C.J.; Caterbi, S.; Franke-Fayard, B.; Bonilla, J.A.; Syphard, L.M.; Di Cristina, M.; Dottorini, T.; Savarino, A.; Cassone, A.; et al. Plasmepsin 4-Deficient Plasmodium berghei Are Virulence Attenuated and Induce Protective Immunity against Experimental Malaria. Am. J. Pathol. 2010, 176, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Francisco, J.S.; Barría, I.; Gutiérrez, B.; Neira, I.; Muñoz, C.; Sagua, H.; Araya, J.E.; Andrade, J.C.; Zailberger, A.; Catalán, A.; et al. Decreased cruzipain and gp85/trans-sialidase family protein expression contributes to loss of Trypanosoma cruzi trypomastigote virulence. Microbes Infect. 2017, 19, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Wright, I.G.; Goodger, B.V.; Mahoney, D.F. Virulent and Avirulent Strains ofBabesia bovis: The Relationship Between Parasite Protease Content and Pathophysiological Effect of the Strain. J. Protozool. 1981, 28, 118–120. [Google Scholar] [CrossRef]
- Savon, L.C.; Alonso, M.; Rodriguez-Diego, J.; Blandino, T. Determination of the protease activity in a Cuban strain of Babesia bovis. Rev. Elev. Med. Vet. Pays Trop. 1992, 45, 30–31. [Google Scholar] [CrossRef]
- Adamson, R.E.; Hall, F.R. Matrix metalloproteinases mediate the metastatic phenotype of Theileria annulata-transformed cells. Parasitology 1996, 113, 449–455. [Google Scholar] [CrossRef]
- Adamson, R.; Logan, M.; Kinnaird, J.; Langsley, G.; Hall, R. Loss of matrix metalloproteinase 9 activity in Theileria annulata-attenuated cells is at the transcriptional level and is associated with differentially expressed AP-1 species. Mol. Biochem. Parasitol. 2000, 106, 51–61. [Google Scholar] [CrossRef]
- Shkap, V.; Pipano, E.; Rasulov, I.; Azimov, D.; Savitsky, I.; Fish, L.; Krigel, Y.; Leibovitch, B. Proteolytic enzyme activity and attenuation of virulence in Theileria annulata schizont-infected cells. Vet. Parasitol. 2003, 115, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.M.; Beyer, D.; Bakheit, M.A.; Kullmann, B.; Salih, D.A.; Ahmed, J.S.; Seitzer, U. Influence of subculturing on gene expression in a Theileria lestoquardi-infected cell line. Vaccine 2008, 26 (Suppl. 6), G17–G23. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Guan, G.; Liu, J.; Liu, A.; Li, Y.; Yin, H.; Luo, J. Screening and identification of host proteins interacting with Theileria annulata cysteine proteinase (TaCP) by yeast-two-hybrid system. Parasit. Vectors 2017, 10, 536. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Waller, M.; Barrett, A.J.; Bateman, A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014, 42, D503–D509. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Col-well, L.; et al. InterPro in 2022. Nucleic Acids Res. 2022, 51, D418–D427. [Google Scholar] [CrossRef] [PubMed]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Armenteros, J.J.A.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv 2022. [Google Scholar]
- Tatusov, R.L.; Koonin, E.V.; Lipman, D.J. A Genomic Perspective on Protein Families. Science 1997, 278, 631–637. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]



| Proteinases and Nonproteinase Homologs | |||||||
|---|---|---|---|---|---|---|---|
| Proteinase Repertoire | Total | Proteinases vs. Nonproteinase Homologs a | Catalytic Type | ||||
| Aspartic | Cysteine | Metallo | Serine | Threonine | |||
| B. bovis | 133 | 82/51 | 6 | 26 | 36 | 51 | 14 |
| T. annulata | 132 | 80/52 | 8 | 36 | 35 | 38 | 15 |
| Proteinases and Nonproteinase Homologs a | |||||||
|---|---|---|---|---|---|---|---|
| Total | Proteinases vs. Nonproteinase Homologs | Catalytic Type | |||||
| Aspartic | Cysteine | Metallo | Serine | Threonine | |||
| (a) Orthologous proteinases | |||||||
| Orthologous pairs | 108 | 5 | 24 | 32 | 33 | 14 | |
| Proteinases vs. non proteinase homologs | 108 | 54/48/6 a | 5/0 | 18/6 | 15/17 | 11/22 | 5/9 |
| (b) Species-specific proteinases | |||||||
| B. bovis-specific | 25 | 7/18 | 1 | 2 | 4 | 18 | 0 |
| T. annulata-specific | 24 | 14/10 | 3 | 12 | 3 | 5 | 1 |
| Dual Domain Composition | Gene ID | Upstream Domain a | Downstream Domain b |
|---|---|---|---|
| M16-M16 | BBOV_III003850/TA11975 | a nf | b nf |
| BBOV_IV001260/TA19130 | a f | b nf | |
| M23-M23 | BBOV_II001730 | a nf | b nf |
| S9-S9 | BBOV_III011180 | a nf | b nf |
| BBOV_IV003330 | a nf | b nf | |
| BBOV_II003080 | a nf | b nf |
| Catalytic Type | Family a | B. bovis | T. annulata | ||
|---|---|---|---|---|---|
| Gene ID b | n | Gene ID b | n | ||
| Aspartic | A1 | BBOV_IV010360 | 1 | - | 0 |
| Cysteine | C1 | - | 0 | TA03720; TA03735; TA03740;TA03745; TA03750; TA10955; TA15660; TA15715 | 8 |
| C12 | BBOV_III010630 | 1 | - | 0 | |
| C19 | - | 0 | TA16070 | 1 | |
| C48 | - | 0 | TA11130 | 1 | |
| C86 | - | 0 | TA12030 | 1 | |
| C88 | - | 0 | TA14855 | 1 | |
| C97 | BBOV_II001800 | 1 | - | 0 | |
| Metallo | M16 | - | 0 | TA12175 | 1 |
| Serine | S8 | BBOV_II006080 | 1 | - | 0 |
| S9 | BBOV_IV005690 | 1 | - | 0 | |
| S33 | - | 0 | TA18005 | 1 | |
| S54 | BBOV_II005930; BBOV_II005940; BBOV_II006070; BBOV_III008600 | 4 | - | 1 | |
| Threonine | T1 | - | 0 | TA15900 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poklepovich, T.J.; Mesplet, M.; Gallenti, R.; Florin-Christensen, M.; Schnittger, L. Comparative Degradome Analysis of the Bovine Piroplasmid Pathogens Babesia bovis and Theileria annulata. Pathogens 2023, 12, 237. https://doi.org/10.3390/pathogens12020237
Poklepovich TJ, Mesplet M, Gallenti R, Florin-Christensen M, Schnittger L. Comparative Degradome Analysis of the Bovine Piroplasmid Pathogens Babesia bovis and Theileria annulata. Pathogens. 2023; 12(2):237. https://doi.org/10.3390/pathogens12020237
Chicago/Turabian StylePoklepovich, Tomás Javier, Maria Mesplet, Romina Gallenti, Monica Florin-Christensen, and Leonhard Schnittger. 2023. "Comparative Degradome Analysis of the Bovine Piroplasmid Pathogens Babesia bovis and Theileria annulata" Pathogens 12, no. 2: 237. https://doi.org/10.3390/pathogens12020237