Upf2-Mediated Nonsense-Mediated Degradation Pathway Involved in Genetic Compensation of TrpA1 Knockout Mutant Silkworm (Bombyx mori)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Silkworm, B. mori
2.2. Construction of the BmTrpA1−/− Mutant and Prediction of the Three-Dimensional Structure of the Protein
2.3. Mutant Screening
2.4. Investigation of Larval Growth and Development and Cocoon Economic Traits
2.5. Extraction of Total RNA from Silkworm Tissues and RT-qPCR Analysis
2.6. Embryonic RNA Interference
3. Results
3.1. Construction of a BmTrpA1−/− Mutant Strain
3.2. No Obvious Differences Were Observed in the Growth, Development, or Diapause Phenotype between BmTrpA1−/− and wt
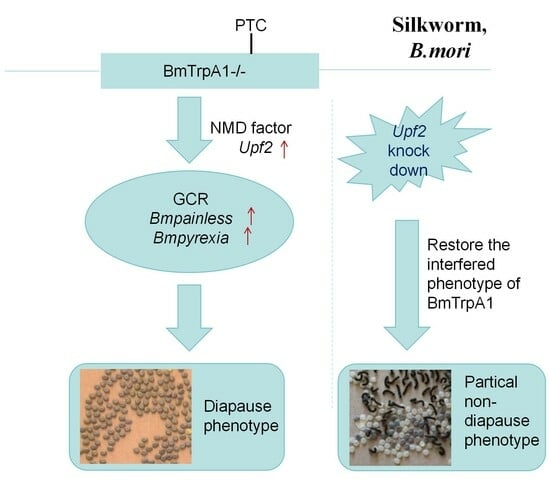
3.3. BmPyrexia, BmPainless, and BmUpf2 Showed Upregulated Expression in BmTrpA1−/− Mutant
3.4. RNA Interference of the UPF2 Gene Alters the Mutant’s Diapause Phenotype
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jakutis, G.; Stainier, D.Y. Genotype–Phenotype relationships in the context of transcriptional adaptation and genetic robustness. Annu. Rev. Genet. 2021, 55, 71–91. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, Y.; Zhang, D.; Dai, X.; Estelle, M.; Zhao, Y. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc. Natl. Acad. Sci. USA 2015, 112, 2275–2280. [Google Scholar] [CrossRef] [PubMed]
- De Souza, A.T.; Dai, X.; Spencer, A.G.; Reppen, T.; Menzie, A.; Roesch, P.L.; He, Y.; Caguyong, M.J.; Bloomer, S.; Herweijer, H. Transcriptional and phenotypic comparisons of Ppara knockout and siRNA knockdown mice. Nucleic Acids Res. 2006, 34, 4486–4494. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Jaiswal, M.; Charng, W.-L.; Gambin, T.; Karaca, E.; Mirzaa, G.; Wiszniewski, W.; Sandoval, H.; Haelterman, N.A.; Xiong, B. A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell 2014, 159, 200–214. [Google Scholar] [CrossRef] [PubMed]
- Kok, F.O.; Shin, M.; Ni, C.-W.; Gupta, A.; Grosse, A.S.; van Impel, A.; Kirchmaier, B.C.; Peterson-Maduro, J.; Kourkoulis, G.; Male, I. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell 2015, 32, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Morgens, D.W.; Deans, R.M.; Li, A.; Bassik, M.C. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol. 2016, 34, 634–636. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-F.; Imam, J.S.; Wilkinson, M.F. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007, 76, 51–74. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Panigrahi, G.K. Messenger RNA Surveillance: Current Understanding, Regulatory Mechanisms, and Future Implications. Mol. Biotechnol. 2024, 1–17. [Google Scholar] [CrossRef]
- El-Brolosy, M.A.; Kontarakis, Z.; Rossi, A.; Kuenne, C.; Günther, S.; Fukuda, N.; Kikhi, K.; Boezio, G.L.; Takacs, C.M.; Lai, S.-L. Genetic compensation triggered by mutant mRNA degradation. Nature 2019, 568, 193–197. [Google Scholar] [CrossRef]
- Ma, Z.; Zhu, P.; Shi, H.; Guo, L.; Zhang, Q.; Chen, Y.; Chen, S.; Zhang, Z.; Peng, J.; Chen, J. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature 2019, 568, 259–263. [Google Scholar] [CrossRef]
- Xie, A.; Ma, Z.; Wang, J.; Zhang, Y.; Chen, Y.; Yang, C.; Chen, J.; Peng, J. Upf3a but not Upf1 mediates the genetic compensation response induced by leg1 deleterious mutations in an H3K4me3-independent manner. Cell Discov. 2023, 9, 63. [Google Scholar] [CrossRef] [PubMed]
- Meents, J.E.; Ciotu, C.I.; Fischer, M.J. TRPA1: A molecular view. J. Neurophysiol. 2019, 121, 427–443. [Google Scholar] [CrossRef]
- Paulsen, C.E.; Armache, J.-P.; Gao, Y.; Cheng, Y.; Julius, D. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature 2015, 520, 511–517. [Google Scholar] [CrossRef]
- Matsuura, H.; Sokabe, T.; Kohno, K.; Tominaga, M.; Kadowaki, T. Evolutionary conservation and changes in insect TRP channels. BMC Evol. Biol. 2009, 9, 228. [Google Scholar] [CrossRef]
- Lee, Y.; Lee, Y.; Lee, J.; Bang, S.; Hyun, S.; Kang, J.; Hong, S.-T.; Bae, E.; Kaang, B.-K.; Kim, J. Pyrexia is a new thermal transient receptor potential channel endowing tolerance to high temperatures in Drosophila melanogaster. Nat. Genet. 2005, 37, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, M.; Caterina, M.J. Thermosensation and pain. J. Neurobiol. 2004, 61, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Tracey, W.D.; Wilson, R.I.; Laurent, G.; Benzer, S. painless, a Drosophila gene essential for nociception. Cell 2003, 113, 261–273. [Google Scholar] [CrossRef]
- Castillo, K.; Diaz-Franulic, I.; Canan, J.; Gonzalez-Nilo, F.; Latorre, R. Thermally activated TRP channels: Molecular sensors for temperature detection. Phys. Biol. 2018, 15, 021001. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Sokabe, T.; Kashio, M.; Yasukochi, Y.; Tominaga, M.; Shiomi, K. Embryonic thermosensitive TRPA1 determines transgenerational diapause phenotype of the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. USA 2014, 111, E1249–E1255. [Google Scholar] [CrossRef]
- Huang, Z. A Novel Regulatory Mechanism Underlying the Regulation of Ambient Temperature on Larval Growth and Development in Silkworm. Master’s Thesis, Southwest University, Chongqing, China, 2023. [Google Scholar]
- Chen, Y.-r.; Jiang, T.; Zhu, J.; Xie, Y.-c.; Tan, Z.-c.; Chen, Y.-h.; Tang, S.-m.; Hao, B.-f.; Wang, S.-p.; Huang, J.-s. Transcriptome sequencing reveals potential mechanisms of diapause preparation in bivoltine silkworm Bombyx mori (Lepidoptera: Bombycidae). Comp. Biochem. Physiol. Part D Genom. Proteom. 2017, 24, 68–78. [Google Scholar]
- Zhu, J.; Chen, Y.-R.; Geng, T.; Tang, S.-M.; Zhao, Q.-l.; Shen, X.-J. A 14-amino acids deletion in BmShadow results to non-moult on the 2nd instar in the bivoltine silkworm, Bombyx mori. Gene 2021, 777, 145450. [Google Scholar] [CrossRef] [PubMed]
- Bienert, S.; Waterhouse, A.; De Beer, T.A.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository—New features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, X.; Shiotsuki, T.; Wang, Z.; Xu, X.; Huang, Y.; Li, M.; Li, K.; Tan, A. Depletion of juvenile hormone esterase extends larval growth in Bombyx mori. Insect Biochem. Mol. Biol. 2017, 81, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Masumoto, M.; Yaginuma, T.; Niimi, T. Functional analysis of Ultrabithorax in the silkworm, Bombyx mori, using RNAi. Dev. Genes Evol. 2009, 219, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Tian, Y.; Peng, Y.; Zheng, S. Knock down of target genes by RNA interference in the embryos of lepidopteran insect, Bombyx mori. STAR Protoc. 2022, 3, 101219. [Google Scholar] [CrossRef]
- Tsuchiya, R.; Kaneshima, A.; Kobayashi, M.; Yamazaki, M.; Takasu, Y.; Sezutsu, H.; Tanaka, Y.; Mizoguchi, A.; Shiomi, K. Maternal GABAergic and GnRH/corazonin pathway modulates egg diapause phenotype of the silkworm Bombyx mori. Proc. Natl. Acad. Sci. USA 2021, 118, e2020028118. [Google Scholar] [CrossRef]
- Clerici, M.; Deniaud, A.; Boehm, V.; Gehring, N.H.; Schaffitzel, C.; Cusack, S. Structural and functional analysis of the three MIF4G domains of nonsense-mediated decay factor UPF2. Nucleic Acids Res. 2014, 42, 2673–2686. [Google Scholar] [CrossRef] [PubMed]
- Colón, E.M.; Haddock, L.A., III; Lasalde, C.; Lin, Q.; Ramírez-Lugo, J.S.; González, C.I. Characterization of the mIF4G Domains in the RNA Surveillance Protein Upf2p. Curr. Issues Mol. Biol. 2023, 46, 244–261. [Google Scholar] [CrossRef]





| Primer Name | Primer Sequence (5′–3′) | Primer Purpose |
|---|---|---|
| TrpA1-582-F | GATAACCGATCTGGACGCTGAATG | Mutation site sequencing detection |
| TrpA1-880-R | CTTGAGCGCAAGCTAAGTGCACAG | |
| gRNA-771-793 | TtctaatacgactcactatagCGTTCATGGAGGTGATA TCAgttttagagctaga | Synthesis of sgRNA in vitro |
| Primer Name | Primer Sequence (5′–3′) | Gene Accession No. |
|---|---|---|
| BmPainless-F | CGGTCTTGCGGTTAGTGACA | NM_001309624.1 |
| BmPainless-R | GCTACCGATAAGCACGCTCT | |
| BmPyrexia-F | ATGATGGCCGCTTACGACAT | NM_001309607.1 |
| BmPyrexia-R | TCCGAGTCCTGAGTAACCGT | |
| BmUpf1-F | GCGAGAGGCAATGGAGTCTT | XM_004929604.4 |
| BmUpf1-R | CACCGGCTTCTTCCTTGAGT | |
| BmUpf2-F | CATTGCTGTCCCGATGACCT | XM_038010585.1 |
| BmUpf2-R | AACGAATTCCACGCCCTCTT | |
| BmUpf3a-F | GGATCGGAAGAGACAGACACA | NM_001046855.2 |
| BmUpf3a-R | TTCTTTCGCGAGACGCTGTT | |
| BmActin3-F | CGGCTACTCGTTCACTACC | NM_001126254.1 |
| BmActin3-R | CCGTCGGGAAGTTCGTAAG | |
| BmRp49-F | TCAATCGGATCGCTATGACA | NM_001098282.2 |
| BmRp49-R | ATGACGGGTCTTCTTGTTGG |
| Primer Name | Primer Sequence (5′–3′) |
|---|---|
| dsGFP-F | ACGTAAACGGCCACAAGTTC |
| T7dsGFP-F | TAATACGACTCACTATAGGGACGTAAACGGCCACAAGTTC |
| dsGFP-R | TGTTCTGCTGGTAGTGGTCG |
| T7dsGFP-R | TAATACGACTCACTATAGGGTGTTCTGCTGGTAGTGGTCG |
| dsUpf1-F | CTCGCAATCGCTCACGTTTC |
| T7dsUpf1-F | TAATACGACTCACTATAGGGCTCGCAATCGCTCACGTTTC |
| dsUpf1-R | ACATTCCGAGCACCACATGA |
| T7dsUpf1-R | TAATACGACTCACTATAGGGACATTCCGAGCACCACATGA |
| DsUpf2-F | TCATCAAAACTGCGGGTGGAT |
| T7dsUpf2-F | TAATACGACTCACTATAGGGTCATCAAAACTGCGGGTGGAT |
| DsUpf2-R | TGAGAGTTCTCCTCTGGTGTG |
| T7dsUpf2-R | TAATACGACTCACTATAGGGTGAGAGTTCTCCTCTGGTGTG |
| Group | Whole Cocoon Weight of Females (g) | Whole Cocoon Weight of Males (g) | Cocoon Shell Weight of Females (g) | Cocoon Shell Weight of Males (g) |
|---|---|---|---|---|
| WT (n = 30) | 1.673 ± 0.1385 | 1.300 ± 0.06543 | 0.3189 ± 0.03326 | 0.2973 ± 0.03197 |
| TRPA1−/−(n = 30) | 1.744 ± 0.1559 | 1.479 ± 0.1320 | 0.3014 ± 0.02375 | 0.299 ± 0.02042 |
| p Value | 0.0683 | <0.0001 | 0.0225 | 0.8145 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.-Y.; Zhu, J.; Zhang, Y.-Z.; Cui, Q.-Y.; Wang, S.-S.; Ning, Y.-W.; Shen, X.-J. Upf2-Mediated Nonsense-Mediated Degradation Pathway Involved in Genetic Compensation of TrpA1 Knockout Mutant Silkworm (Bombyx mori). Insects 2024, 15, 313. https://doi.org/10.3390/insects15050313
Wang D-Y, Zhu J, Zhang Y-Z, Cui Q-Y, Wang S-S, Ning Y-W, Shen X-J. Upf2-Mediated Nonsense-Mediated Degradation Pathway Involved in Genetic Compensation of TrpA1 Knockout Mutant Silkworm (Bombyx mori). Insects. 2024; 15(5):313. https://doi.org/10.3390/insects15050313
Chicago/Turabian StyleWang, Dong-Yue, Juan Zhu, Yi-Zhong Zhang, Qian-Yi Cui, Shan-Shan Wang, Yang-Wei Ning, and Xing-Jia Shen. 2024. "Upf2-Mediated Nonsense-Mediated Degradation Pathway Involved in Genetic Compensation of TrpA1 Knockout Mutant Silkworm (Bombyx mori)" Insects 15, no. 5: 313. https://doi.org/10.3390/insects15050313