DNA Metabarcoding as a Tool for Disentangling Food Webs in Agroecosystems
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Sampling
2.2. DNA Extraction
2.3. PCR and Illumina Sequencing
2.4. Sequence Analysis
3. Results
3.1. Sequence Analyses
3.2. Diet Composition of Predatory Arthropods
3.3. Diet Composition of Insectivorous Vertebrates
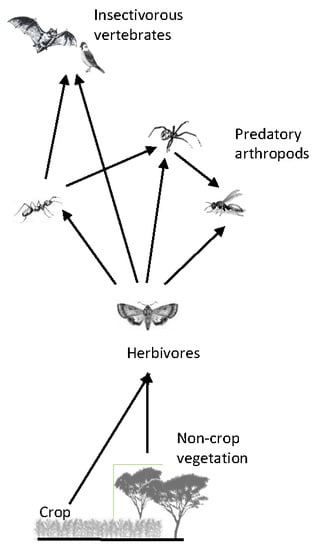
3.4. Qualitative Trophic Network and Interactions
4. Discussion
4.1. Taxonomic Affiliation and Diet Analysis
4.2. Diet Composition of Predatory Arthropods
4.3. Diet Composition of Insectivorous Vertebrates
4.4. Qualitative Trophic Network and Interactions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maxmen, A. Crop pests: Under attack. Nature 2013, 501, S15–S17. [Google Scholar] [CrossRef] [PubMed]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Begg, G.S.; Cook, S.M.; Dye, R.; Ferrante, M.; Franck, P.; Lavigne, C.; Lövei, G.L.; Mansion-Vaquie, A.; Pell, J.K.; Petit, S.; et al. A functional overview of conservation biological control. Crop Prot. 2017, 97, 145–158. [Google Scholar] [CrossRef]
- Gurr, G.M.; Van Emden, H.F.; Wratten, S.D. Habitat manipulation and natural enemy efficiency: implications for the control of pests. In Conservation biological control; Elsevier: San Diego, CA, USA, 1998; pp. 155–183. [Google Scholar]
- Wyckhuys, K.A.G.; Lu, Y.; Morales, H.; Vazquez, L.L.; Legaspi, J.C.; Eliopoulos, P.A.; Hernandez, L.M. Current status and potential of conservation biological control for agriculture in the developing world. Biol. Control 2013, 65, 152–167. [Google Scholar] [CrossRef]
- Symondson, W.O.C.; Sunderland, K.D.; Greenstone, M.H. Can Generalist Predators Be Effective Biocontrol Agents? Annu. Rev. Entomol. 2002, 47, 561–594. [Google Scholar] [CrossRef] [Green Version]
- Harwood, J.D.; Obrycki, J.J. The role of alternative prey in sustaining predator populations. In Proceedings of the Second International Symposium on Biological Control of Arthropods, Davos, Switzerland, 12–16 September 2005; pp. 453–462. [Google Scholar]
- Harwood, J.D.; Desneux, N.; Yoo, H.J.S.; Rowley, D.L.; Greenstone, M.H.; Obrycki, J.J.; O′Neil, R.J. Tracking the role of alternative prey in soybean aphid predation by Orius insidiosus: A molecular approach. Mol. Ecol. 2007, 16, 4390–4400. [Google Scholar] [CrossRef]
- Furlong, M.J. Knowing your enemies: Integrating molecular and ecological methods to assess the impact of arthropod predators on crop pests. Insect Sci. 2015, 22, 6–19. [Google Scholar] [CrossRef]
- Jonsson, M.; Wratten, S.D.; Landis, D.A.; Gurr, G.M. Recent advances in conservation biological control of arthropods by arthropods. Biol. Control 2008, 45, 172–175. [Google Scholar] [CrossRef]
- Pompanon, F.; Deagle, B.E.; Symondson, W.O.C.; Brown, D.S.; Jarman, S.N.; Taberlet, P. Who is eating what: Diet assessment using next generation sequencing. Mol. Ecol. 2012, 21, 1931–1950. [Google Scholar] [CrossRef] [Green Version]
- Waldner, T.; Traugott, M. DNA-based analysis of regurgitates: A noninvasive approach to examine the diet of invertebrate consumers. Mol. Ecol. Resour. 2012, 12, 669–675. [Google Scholar] [CrossRef]
- Bohmann, K.; Monadjem, A.; Noer, C.L.; Rasmussen, M.; Zeale, M.R.K.; Clare, E.; Jones, G.; Willerslev, E.; Gilbert, M.T.P. Molecular Diet Analysis of Two African Free-Tailed Bats (Molossidae) Using High Throughput Sequencing. PLoS ONE 2011, 6, e21441. [Google Scholar] [CrossRef] [PubMed]
- Galan, M.; Pons, J.-B.; Tournayre, O.; Pierre, É.; Leuchtmann, M.; Pontier, D.; Charbonnel, N. Metabarcoding for the parallel identification of several hundred predators and their prey: Application to bat species diet analysis. Mol. Ecol. Resour. 2018, 18, 474–489. [Google Scholar] [CrossRef] [PubMed]
- Paula, D.P.; Linard, B.; Crampton-Platt, A.; Srivathsan, A.; Timmermans, M.J.T.N.; Sujii, E.R.; Pires, C.S.S.; Souza, L.M.; Andow, D.A.; Vogler, A.P. Uncovering Trophic Interactions in Arthropod Predators through DNA Shotgun-Sequencing of Gut Contents. PLoS ONE 2016, 11, e0161841. [Google Scholar] [CrossRef] [Green Version]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Brochmann, C.; Willerslev, E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 2012, 21, 2045–2050. [Google Scholar] [CrossRef] [PubMed]
- Firlej, A.; Doyon, J.; Harwood, J.D.; Brodeur, J. A Multi-Approach Study to Delineate Interactions Between Carabid Beetles and Soybean Aphids. Environ. Entomol. 2013, 42, 89–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kartzinel, T.R.; Chen, P.A.; Coverdale, T.C.; Erickson, D.L.; Kress, W.J.; Kuzmina, M.L.; Rubenstein, D.I.; Wang, W.; Pringle, R.M. DNA metabarcoding illuminates dietary niche partitioning by African large herbivores. Proc. Natl. Acad. Sci. USA 2015, 112, 8019–8024. [Google Scholar] [CrossRef] [Green Version]
- Lopes, C.M.; De Barba, M.; Boyer, F.; Mercier, C.; da Silva Filho, P.J.S.; Heidtmann, L.M.; Galiano, D.; Kubiak, B.B.; Langone, P.; Garcias, F.M. DNA metabarcoding diet analysis for species with parapatric vs. sympatric distribution: A case study on subterranean rodents. Heredity 2015, 114, 525. [Google Scholar] [CrossRef] [Green Version]
- Mollot, G.; Duyck, P.-F.; Lefeuvre, P.; Lescourret, F.; Martin, J.-F.; Piry, S.; Canard, E.; Tixier, P. Cover Cropping Alters the Diet of Arthropods in a Banana Plantation: A Metabarcoding Approach. PLoS ONE 2014, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Greenstone, M.H.; Payton, M.E.; Weber, D.C.; Simmons, A.M. The detectability half-life in arthropod predator–prey research: What it is, why we need it, how to measure it, and how to use it. Mol. Ecol. 2014, 23, 3799–3813. [Google Scholar] [CrossRef]
- Symondson, W.O.; Harwood, J.D. Special issue on molecular detection of trophic interactions: Unpicking the tangled bank. Mol. Ecol. 2014, 23, 3601–3604. [Google Scholar] [CrossRef] [Green Version]
- Bhatnagar, V.S. Conservation and encouragement of natural enemies of insect pests in dryland subsistence farming: Problems, progress and prospects in the Sahelian zone. Int. J. Trop. Insect Sci. 1987, 8, 791–795. [Google Scholar] [CrossRef]
- Gahukar, R.T. Insect pests of earl millet in West Africa. Trop. Pest Manag. 1984, 30, 142–147. [Google Scholar] [CrossRef]
- Soti, V.; Thiaw, I.; Debaly, Z.M.; Sow, A.; Diaw, M.; Fofana, S.; Diakhate, M.; Thiaw, C.; Brévault, T. Effect of landscape diversity and crop management on the control of the millet head miner, Heliocheilus albipunctella (Lepidoptera: Noctuidae) by natural enemies. Biol. Control 2019, 129, 115–122. [Google Scholar] [CrossRef]
- Sow, A.; Seye, D.; Faye, E.; Benoit, L.; Galan, M.; Haran, J.; Brévault, T. Birds and bats contribute to natural regulation of the millet head miner in tree-crop agroforestry systems. Crop Prot. 2020, 105127. [Google Scholar] [CrossRef]
- Sow, A.; Brévault, T.; Benoit, L.; Chapuis, M.-P.; Galan, M.; Coeur D’acier, A.; Delvare, G.; Sembène, M.; Haran, J. Deciphering host-parasitoid interactions and parasitism rates of crop pests using DNA metabarcoding. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sow, A.; Brévault, T.; Delvare, G.; Haran, J.; Benoit, L.; Coeur d’Acier, A.; Galan, M.; Thiaw, C.; Soti, V.; Sembène, M. DNA sequencing to help identify crop pests and their natural enemies in agro–ecosystems: The case of the millet head miner Heliocheilus albipunctella (Lepidoptera: Noctuidae) in sub-Saharan Africa. Biol. Control 2018, 121, 199–207. [Google Scholar] [CrossRef]
- King, R.A.; Read, D.S.; Traugott, M.; Symondson, W.O.C. INVITED REVIEW: Molecular analysis of predation: A review of best practice for DNA-based approaches. Mol. Ecol. 2008, 17, 947–963. [Google Scholar] [CrossRef]
- Pons, J. DNA-based identification of preys from non-destructive, total DNA extractions of predators using arthropod universal primers. Mol. Ecol. Notes 2006, 6, 623–626. [Google Scholar] [CrossRef]
- Zarzoso-Lacoste, D.; Jan, P.-L.; Lehnen, L.; Girard, T.; Besnard, A.-L.; Puechmaille, S.J.; Petit, E.J. Combining noninvasive genetics and a new mammalian sex-linked marker provides new tools to investigate population size, structure and individual behaviour: An application to bats. Mol. Ecol. Resour. 2018, 18, 217–228. [Google Scholar] [CrossRef] [Green Version]
- Gillet, F.; Tiouchichine, M.-L.; Galan, M.; Blanc, F.; Némoz, M.; Aulagnier, S.; Michaux, J.R. A new method to identify the endangered Pyrenean desman (Galemys pyrenaicus) and to study its diet, using next generation sequencing from faeces. Mamm. Biol. Z. Für Säugetierkd. 2015, 80, 505–509. [Google Scholar] [CrossRef]
- Corse, E.; Tougard, C.; Archambaud-Suard, G.; Agnèse, J.-F.; Messu Mandeng, F.D.; Bilong Bilong, C.F.; Duneau, D.; Zinger, L.; Chappaz, R.; Xu, C.C. One-locus-several-primers: A strategy to improve the taxonomic and haplotypic coverage in diet metabarcoding studies. Ecol. Evol. 2019, 9, 4603–4620. [Google Scholar] [CrossRef]
- Tournayre, O.; Leuchtmann, M.; Filippi-Codaccioni, O.; Trillat, M.; Piry, S.; Pontier, D.; Charbonnel, N.; Galan, M. In silico and empirical evaluation of twelve COI & 16S metabarcoding primer sets for insectivorous diet analyses. bioRxiv 2019, 742874. [Google Scholar] [CrossRef]
- Galan, M.; Razzauti, M.; Bard, E.; Bernard, M.; Brouat, C.; Charbonnel, N.; Dehne-Garcia, A.; Loiseau, A.; Tatard, C.; Tamisier, L.; et al. 16S rRNA Amplicon Sequencing for Epidemiological Surveys of Bacteria in Wildlife. mSystems 2016, 1, e00032-16. [Google Scholar] [CrossRef] [Green Version]
- Robasky, K.; Lewis, N.E.; Church, G.M. The role of replicates for error mitigation in next-generation sequencing. Nat. Rev. Genet. 2014, 15, 56–62. [Google Scholar] [CrossRef]
- Sow, A.; Haran, J.; Benoit, L.; Galan, M.; Brévault, T. DNA Metabarcoding as a Tool for Disentangling Food Webs in Agroecosystems. Available online: https://zenodo.org/record/3752026#.XrZMDJl5tPY (accessed on 14 April 2020).
- Escudié, F.; Auer, L.; Bernard, M.; Mariadassou, M.; Cauquil, L.; Vidal, K.; Maman, S.; Hernandez-Raquet, G.; Combes, S.; Pascal, G. FROGS: Find, Rapidly, OTUs with Galaxy Solution. Bioinformatics 2017, 34, 1287–1294. [Google Scholar] [CrossRef] [PubMed]
- Sow, A.; Brévault, T.; Benoit, L.; Chapuis, M.-P.; Galan, M.; Coeur D’Acier, A.; Delvare, G.; Sembène, M.; Haran, J. Data from: Deciphering host-parasitoid interactions and parasitism rates of crop pests using DNA metabarcoding. Dryad Dataset 2019. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinforma. Oxf. Engl. 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinforma. Oxf. Engl. 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Mahé, F.; Rognes, T.; Quince, C.; de Vargas, C.; Dunthorn, M. Swarm: Robust and fast clustering method for amplicon-based studies. PeerJ 2014, 2, e593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratnasingham, S.; Hebert, P.D. BOLD: The Barcode of Life Data System (www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2016, 44, D67–D72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alberdi, A.; Aizpurua, O.; Gilbert, M.T.P.; Bohmann, K. Scrutinizing key steps for reliable metabarcoding of environmental samples. Methods Ecol. Evol. 2017, 9, 134–147. [Google Scholar] [CrossRef] [Green Version]
- Valentini, A.; Pompanon, F.; Taberlet, P. DNA barcoding for ecologists. Trends Ecol. Evol. 2009, 24, 110–117. [Google Scholar] [CrossRef]
- Corse, E.; Meglécz, E.; Archambaud, G.; Ardisson, M.; Martin, J.-F.; Tougard, C.; Chappaz, R.; Dubut, V. A from-benchtop-to-desktop workflow for validating HTS data and for taxonomic identification in diet metabarcoding studies. Mol. Ecol. Resour. 2017, 17, e146–e159. [Google Scholar] [CrossRef]
- Gagnon, A.-È.; Doyon, J.; Heimpel, G.E.; Brodeur, J. Prey DNA detection success following digestion by intraguild predators: Influence of prey and predator species. Mol. Ecol. Resour. 2011, 11, 1022–1032. [Google Scholar] [CrossRef]
- Greenstone, M.H.; Weber, D.C.; Coudron, T.A.; Payton, M.E.; Hu, J.S. Removing external DNA contamination from arthropod predators destined for molecular gut-content analysis. Mol. Ecol. Resour. 2012, 12, 464–469. [Google Scholar] [CrossRef]
- Boukary, I.B.; Tourneur, J.-C.; Gingras, J. Life cycle of Forficula Senegalensis Serv. (Dermaptera: Forficulidae) and its relationship to the development of bulrush millet in the Sudanese-Sahelian zone of Niger. Can. Entomol. 1996, 128, 831–838. [Google Scholar] [CrossRef]
- Cotes, B.; González, M.; Benítez, E.; De Mas, E.; Clemente-Orta, G.; Campos, M.; Rodríguez, E. Spider communities and biological control in native habitats surrounding greenhouses. Insects 2018, 9, 33. [Google Scholar] [CrossRef] [Green Version]
- Pfannenstiel, R.S. Spider predators of lepidopteran eggs in south Texas field crops. Biol. Control 2008, 46, 202–208. [Google Scholar] [CrossRef]
- Quan, X.; Wu, L.; Zhou, Q.; Yun, Y.; Peng, Y.; Chen, J. Identification of predation by spiders on the diamondback moth Plutella xylostella. Bull. Insectology 2011, 64, 223–227. [Google Scholar]
- Hanson, H.I.; Palmu, E.; Birkhofer, K.; Smith, H.G.; Hedlund, K. Agricultural Land Use Determines the Trait Composition of Ground Beetle Communities. PLOS ONE 2016, 11, e0146329. [Google Scholar] [CrossRef] [PubMed]
- King, R.A.; Vaughan, I.P.; Bell, J.R.; Bohan, D.A.; Symondson, W.O.C. Prey choice by carabid beetles feeding on an earthworm community analysed using species- and lineage-specific PCR primers. Mol. Ecol. 2010, 19, 1721–1732. [Google Scholar] [CrossRef] [PubMed]
- Kromp, B. Carabid beetles in sustainable agriculture: A review on pest control efficacy, cultivation impacts and enhancement. Agric. Ecosyst. Environ. 1999, 74, 187–228. [Google Scholar] [CrossRef]
- Hernandez, L.M.; Stonedahl, G.M. A review of the economically important species of the genus Orius (Heteroptera: Anthocoridae) in East Africa. J. Nat. Hist. 1999, 33, 543–568. [Google Scholar] [CrossRef]
- Philpott, S.M.; Armbrecht, I. Biodiversity in tropical agroforests and the ecological role of ants and ant diversity in predatory function. Ecol. Entomol. 2006, 31, 369–377. [Google Scholar] [CrossRef]
- Vercambre, B. Raghuva spp. et Masalia sp., chenilles des chandelles du mil en zone sahelienne. Agron. Trop. 1978, 33, 62–79. [Google Scholar]
- Maselou, D.A.; Perdikis, D.C.; Sabelis, M.W.; Fantinou, A.A. Use of plant resources by an omnivorous predator and the consequences for effective predation. Biol. Control 2014, 79, 92–100. [Google Scholar] [CrossRef]
- Kingdon, J. East African Mammals: An Atlas of Evolution in Africa; Academic Press: Chicago, IL, USA, 1974. [Google Scholar]
- Brévault, T.; Ndiaye, A.; Badiane, D.; Bal, A.B.; Sembène, M.; Silvie, P.; Haran, J. First records of the fall armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae), in Senegal. Entomol. Gen. 2018, 37, 129–142. [Google Scholar] [CrossRef]
- Durocher-Granger, L.; Rwomushana, I.; Cole, S.; Babendreier, D.; Huesing, J.E.; Jepson, P.C.; Eddy, R.; Prasanna, B.M. Fall armyworm (FAW) on sorghum in Senegal. In Pest management decision guides; CABI: Wallingford, UK, 2018. [Google Scholar]
- Durocher-Granger, L.; Babendreier, D.; Huesing, J.E.; Jepson, P.C.; Eddy, R.; Prasanna, B.M. Fall armyworm (FAW) on maize in Senegal. In Pest management decision guides; CABI: Wallingford, UK, 2018. [Google Scholar]
- Diatte, M.; Brevault, T.; Sylla, S.; Tendeng, E.; Sall-Sy, D.; Diarra, K. Arthropod pest complex and associated damage in field-grown tomato in Senegal. Int. J. Trop. Insect Sci. 2018, 38, 243–253. [Google Scholar] [CrossRef]
- Kunz, T.H.; de Torrez, E.B.; Bauer, D.; Lobova, T.; Fleming, T.H. Ecosystem services provided by bats. Ann. N. Y. Acad. Sci. 2011, 1223, 1–38. [Google Scholar] [CrossRef]
- Dengis, C.A. Taphozous mauritianus. Mamm. Species 1996, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Fenton, M.B. The foraging behaviour and ecology of animal-eating bats. Can. J. Zool. 1990, 68, 411–422. [Google Scholar] [CrossRef]
- Bruggers, R.L.; Jaeger, M.E.; Jaeger, M.M. Tisserins gendarmes (Ploceus cucullatus abyssinicus) et tisserins masqués (Ploceus intermedius intermedius) munis d’émetteurs radio et de rubans dans une colonie de nidification du Sud de l’Ethopie. Oiseaux Rev. Fr. Ornithol. 1985, 55, 81–92. [Google Scholar]
- Collias, N.E.; Collias, E.C. The behaviour of the West African Village Weaver Bird. Ibis 1970, 112, 457–480. [Google Scholar] [CrossRef]
- Finke, D.L.; Denno, R.F. Predator diversity and the functioning of ecosystems: The role of intraguild predation in dampening trophic cascades. Ecol. Lett. 2005, 8, 1299–1306. [Google Scholar] [CrossRef]
- Finke, D.L.; Denno, R.F. Intraguild Predation Diminished in Complex-Structured Vegetation: Implications for Prey Suppression. Ecology 2002, 83, 643–652. [Google Scholar] [CrossRef]
- Mooney, K.A.; Gruner, D.S.; Barber, N.A.; Bael, S.A.V.; Philpott, S.M.; Greenberg, R. Interactions among predators and the cascading effects of vertebrate insectivores on arthropod communities and plants. Proc. Natl. Acad. Sci. USA 2010, 107, 7335–7340. [Google Scholar] [CrossRef] [Green Version]
- Duffy, J.E.; Cardinale, B.J.; France, K.E.; McIntyre, P.B.; Thébault, E.; Loreau, M. The functional role of biodiversity in ecosystems: Incorporating trophic complexity. Ecol. Lett. 2007, 10, 522–538. [Google Scholar] [CrossRef] [Green Version]
- Brévault, T.; Clouvel, P. Pest management: Reconciling farming practices and natural regulations. Crop Prot. 2019, 115, 1–6. [Google Scholar] [CrossRef]




| Predator | Number of Samples Analyzed | Prey | |||
|---|---|---|---|---|---|
| Order (Family) Species | Order | Species | Blast Identity (%) | Positive Samples (%) | |
| Araneae (unidentified) | 12 | Blattodea | Odontotermes sp. | 100 | 8.3 |
| Sp.1 | Coleoptera | Carabidae sp. | 99 | 8.3 | |
| Elateridae sp. | 99 | 25.0 | |||
| Diptera | Rachispoda sp. | 99 | 8.3 | ||
| Lepidoptera | Hypena masurialis | 100 | 8.3 | ||
| Sp.2 | 30 | Blattodea | Odontotermes sp. | 100 | 10.0 |
| Coleoptera | Elateridae sp. | 98 | 3.3 | ||
| Omonadus floralis | 100 | 3.3 | |||
| Hemiptera | Creontiades pallidus | 99 | 3.3 | ||
| Lepidoptera | Hypena masurialis | 100 | 3.3 | ||
| Orthoptera | Calliptamus barbarus | 100 | 3.3 | ||
| Sp.3 | 19 | Coleoptera | Omonadus formicarius | 99 | 5.3 |
| Diptera | Culex sp. | 100 | 5.3 | ||
| Hemiptera | Creontiades pacificus | 100 | 5.3 | ||
| Creontiades pallidus | 99 | 10.5 | |||
| Hymenoptera | Apocryptophagus testaceus | 98 | 5.3 | ||
| Camponotus sp. | 100 | 10.5 | |||
| Ceratosolen fusciceps | 99 | 15.8 | |||
| Coleoptera (Carabidae) | 43 | Araneae | Gnaphosidae sp. | 100 | 2.3 |
| Bradybeanus scalaris | Blattodea | Odontotermes sp. | 100 | 9.3 | |
| Diptera | Atherigona sp. | 97 | 9.3 | ||
| Hymenoptera | Camponotus sp. | 100 | 4.7 | ||
| Ceratosolen fusciceps | 99 | 14.0 | |||
| Encyrtidae sp. | 100 | 4.7 | |||
| Sycophaga testacea | 99 | 4.7 | |||
| Lepidoptera | Aegoceropsis sp. | 100 | 2.3 | ||
| Thysanoptera | Frankliniella schultzei | 100 | 7.0 | ||
| Dermaptera (Forficulidae) | 79 | Hemiptera | Creontiades pallidus | 100 | 1.3 |
| Forficula senegalensis | Lepidoptera | Hypena masurialis | 100 | 1.3 | |
| Orthoptera | Calliptamus barbarus | 100 | 1.3 | ||
| Hemiptera (Pentatomidae) | 13 | Diptera | Cylindromyia bicolor | 100 | 7.7 |
| Carbula curtana | Pegoplata nigroscutellata | 100 | 7.7 | ||
| Entomobryomorpha | Entomobrya ligata | 100 | 7.7 | ||
| Hemiptera (Anthocoridae)Orius maxidentex | 40 | Hemiptera | Creontiades pacificus | 100 | 5.0 |
| Creontiades pallidus | 99 | 7.5 | |||
| Lepidoptera | Amyna axis | 100 | 2.5 | ||
| Ephestia kuehniella | 100 | 2.5 | |||
| Hypena masurialis | 100 | 2.5 | |||
| Masalia nubila | 97 | 2.5 | |||
| Hymenoptera (Formicidae) | 15 | Hemiptera | Carbula sp. | 97 | 6.7 |
| Monomorium sp. | Creontiades pallidus | 99 | 6.7 | ||
| Hymenoptera | Apocryptophagus testaceus | 98 | 6.7 | ||
| Ceratosolen fusciceps | 99 | 6.7 | |||
| Cryptanusia sp. | 98 | 13.3 | |||
| Pachycondyla sp. | 5 | Coleoptera | Bradybaenus scalaris | 100 | 60.0 |
| Lepidoptera | Aegoceropsis sp. | 100 | 20.0 | ||
| Predator | Number of Samples | Prey | |||
|---|---|---|---|---|---|
| Order (Family) Species | Orders Family | Species | Blast Identity (%) | Postive Samples (%) | |
| Chiroptera (Emballonuridae) | 92 | Blattodea | Macrotermes subhyalinus | 100 | 2.17 |
| Taphozous mauritianus | Odontotermes sp. | 100 | 2.72 | ||
| Coleoptera | Philonthus discoideus | 98 | 1.09 | ||
| Diptera | Chrysomya marginalis | 99 | 3.26 | ||
| Hemiptera | Anisops sardeus | 100 | 2.17 | ||
| Campylomma sp. | 100 | 16.30 | |||
| Creontiades pacificus | 100 | 1.09 | |||
| Creontiades pallidus | 99 | 3.99 | |||
| Hymenoptera | Perilampus sp. | 100 | 1.09 | ||
| Lepidoptera | Achyra coelatalis | 99 | 38.04 | ||
| Achyra nudalis | 100 | 13.04 | |||
| Acontia gratiosa | 100 | 3.26 | |||
| Acontia sp. | 100 | 3.26 | |||
| Adisura bella | 100 | 11.41 | |||
| Adoxophyes thoracica | 99 | 3.26 | |||
| Agrius convolvuli | 100 | 3.26 | |||
| Amyna axis | 100 | 9.78 | |||
| Ancylosis nubeculella | 100 | 7.61 | |||
| Anomis flava | 100 | 3.80 | |||
| Antheua simplex | 99 | 3.26 | |||
| Asota heliconia | 98 | 1.63 | |||
| Bastilla angularis | 100 | 1.09 | |||
| Brachynemata sp. | 99 | 1.09 | |||
| Cadra sp. | 100 | 1.09 | |||
| Caryocolum petrophila | 99 | 1.09 | |||
| Cerastis rubricosa | 98 | 2.17 | |||
| Chrysodeixis acuta | 100 | 1.09 | |||
| Condica capensis | 100 | 3.26 | |||
| Coniesta ignefusalis | 100 | 1.09 | |||
| Daphnis nerii | 100 | 1.09 | |||
| Deltote sp. | 98 | 1.09 | |||
| Duponchelia fovealis | 100 | 2.17 | |||
| Ebertidia haderonides | 98 | 1.09 | |||
| Ectopatria sp. | 100 | 1.09 | |||
| Ericeia inangulata | 99 | 2.17 | |||
| Franclemontia interrogans | 99 | 1.09 | |||
| Garella nilotica | 100 | 5.43 | |||
| Grammodes stolida | 100 | 13.04 | |||
| Helicoverpa armigera | 99 | 3.26 | |||
| Helicoverpa assulta | 100 | 15.22 | |||
| Heliocheilus albipunctella | 100 | 7.61 | |||
| Hippotion gracilis | 100 | 2.17 | |||
| Hyles sp. | 100 | 3.26 | |||
| Hypena holophaea | 100 | 1.09 | |||
| Hypena laceratalis | 100 | 4.35 | |||
| Hypena masurialis | 100 | 4.35 | |||
| Isturgia disputaria | 100 | 3.26 | |||
| Isturgia pulinda | 100 | 4.35 | |||
| Lamoria imbella | 100 | 3.26 | |||
| Lepidoptera | 100 | 1.09 | |||
| Leucania commoides | 100 | 1.09 | |||
| Leucania sp. | 98 | 1.09 | |||
| Lophocrama phoennicochlora | 100 | 1.09 | |||
| Marasmia poeyalis | 100 | 4.35 | |||
| Maurilia sp. | 98 | 1.09 | |||
| Morosaphycita oculiferella | 98 | 2.17 | |||
| Mythimna sp. | 100 | 7.61 | |||
| Noctuidae | 100 | 7.61 | |||
| Ortaliella palaestinensis | 97 | 1.09 | |||
| Pasiphila derasata | 100 | 1.09 | |||
| Pericyma sp. | 100 | 1.09 | |||
| Phazaca theclata | 100 | 2.17 | |||
| Polydesma umbricola | 100 | 1.09 | |||
| Polypogon fractalis | 100 | 1.09 | |||
| Psilopleura vittata | 99 | 1.09 | |||
| Pyrausta sp. | 100 | 1.09 | |||
| Rhabdophera exarata | 99 | 2.17 | |||
| Rhesala moestalis | 100 | 2.17 | |||
| Scoliopteryginae sp | 100 | 2.17 | |||
| Scopelodes sericea | 98 | 1.09 | |||
| Scopula adelpharia | 100 | 1.63 | |||
| Sesamia calamistis | 100 | 8.70 | |||
| Sphingomorpha chlorea | 100 | 1.09 | |||
| Spodoptera cilium | 100 | 4.35 | |||
| Spodoptera exigua | 100 | 7.61 | |||
| Spodoptera littoralis | 100 | 1.09 | |||
| Spodoptera sp. | 100 | 2.17 | |||
| Spoladea recurvalis | 100 | 10.87 | |||
| Stegasta sp. | 98 | 1.09 | |||
| Sympis rufibasis | 99 | 1.09 | |||
| Tegostoma sp. | 100 | 8.70 | |||
| Theclinae | 97 | 1.09 | |||
| Thylacoptila paurosema | 100 | 4.35 | |||
| Thysanopyga cermala | 99 | 1.09 | |||
| Traminda neptunaria | 100 | 1.09 | |||
| Trichoplusia ni | 100 | 10.87 | |||
| Trigonodes hyppasia | 100 | 3.26 | |||
| Trisuloides sp. | 99 | 1.09 | |||
| Uraba lugens | 98 | 2.17 | |||
| Vanessa cardui | 100 | 1.09 | |||
| Neuroptera | Chrysoperla sp | 98 | 1.09 | ||
| Mallada signatus | 98 | 4.35 | |||
| Plesiochrysa atalotis | 99 | 1.09 | |||
| Orthoptera | Acheta domesticus | 99 | 2.17 | ||
| Arcotylus longipes | 99 | 1.09 | |||
| Gryllus bimaculatus | 100 | 5.43 | |||
| Gryllus campestris | 98 | 1.09 | |||
| Locusta migratoria | 100 | 1.09 | |||
| Oecanthus pellucens | 99 | 1.09 | |||
| Oedaleus decorus | 99 | 3.73 | |||
| Oedaleus sp. | 98 | 1.09 | |||
| Stenohippus maculifemur | 99 | 1.09 | |||
| Trilophidia conturbata | 99 | 1.45 | |||
| Passeriformes (Ploceidae) | 80 | Coleoptera | Elateridae | 99 | 1.25 |
| Ploceus cucullatus | Dermaptera | Forficula senegalensis | 100 | 6.25 | |
| Diptera | Bryophaenocladius sp. | 100 | 1.25 | ||
| Hemiptera | Carbula sp. | 98 | 1.25 | ||
| Hymenoptera | Pachycondyla sp. | 100 | 2.5 | ||
| Perilampus sp. | 100 | 1.25 | |||
| Pristomerus pallidus | 100 | 1.25 | |||
| Lepidoptera | Heliocheilus albipunctella | 100 | 2.5 | ||
| Lepidoptera | 98 | 1.25 | |||
| Pelopidas mathias | 100 | 2.5 | |||
| Scoliopteryginae sp | 100 | 1.25 | |||
| Sphingomorpha chlorea | 100 | 2.5 | |||
| Spodoptera cilium | 100 | 1.25 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sow, A.; Haran, J.; Benoit, L.; Galan, M.; Brévault, T. DNA Metabarcoding as a Tool for Disentangling Food Webs in Agroecosystems. Insects 2020, 11, 294. https://doi.org/10.3390/insects11050294
Sow A, Haran J, Benoit L, Galan M, Brévault T. DNA Metabarcoding as a Tool for Disentangling Food Webs in Agroecosystems. Insects. 2020; 11(5):294. https://doi.org/10.3390/insects11050294
Chicago/Turabian StyleSow, Ahmadou, Julien Haran, Laure Benoit, Maxime Galan, and Thierry Brévault. 2020. "DNA Metabarcoding as a Tool for Disentangling Food Webs in Agroecosystems" Insects 11, no. 5: 294. https://doi.org/10.3390/insects11050294