Insight into the Functional Diversification of Lipases in the Endoparasitoid Pteromalus puparum (Hymenoptera: Pteromalidae) by Genome-scale Annotation and Expression Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Annotation of Encoding Lipase Genes
2.2. Expression Patterns Analysis
2.3. RNA Extraction and cDNA Synthesis
2.4. RT-qPCR
2.5. Identification of the Catalytic Triads in P. puparum Lipases
2.6. Search for the β9 loop and Lid Motifs in P. puparum Lipases
2.7. Phylogenetic Analysis
3. Results
3.1. Genome-Scale Identification of Encoding Lipases in Parasitoid Wasps
3.2. Expression Patterns of Encoding Lipases in P. puparum
3.3. Incomplete Catalytic Triads in Predicted Lipases of P. puparum
3.4. Identification of β9 Loop and Lids of Lipases in P. puparum
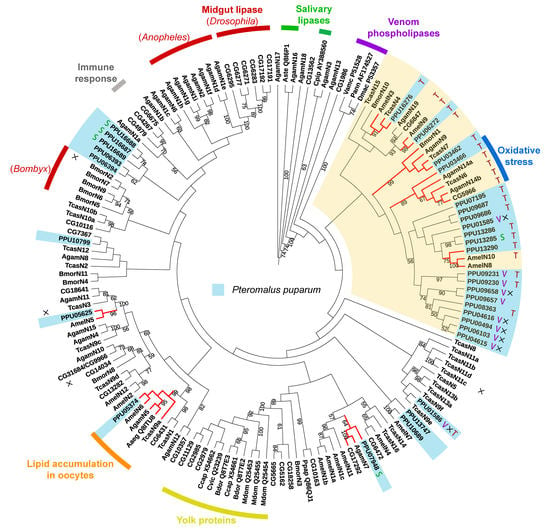
3.5. Phylogenetic Analysis of Lipases in P. puparum
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Derewenda, Z.S. Structure and function of lipases. Adv. Protein Chem. 1994, 45, 1–52. [Google Scholar]
- Holmquist, M. Alpha/beta-hydrolase fold enzymes: Structures, functions and mechanisms. Curr. Protein Pept. Sci. 2000, 1, 209–235. [Google Scholar] [CrossRef]
- Yuan, C.H.; Tsai, M.D. Pancreatic phospholipase A(2): New views on old issues. BBA-Mol. Cell Biol. Lipids 1999, 1441, 215–222. [Google Scholar] [CrossRef]
- Carriere, F.; Withers-Martinez, C.; van Tilberugh, H.; Roussel, A.; Cambillau, C.; Verger, R. Structural basis for the substrate selectivity of pancreatic lipases and some related proteins. BBA-Rev. Biomembr. 1998, 1376, 417–432. [Google Scholar] [CrossRef]
- Gargouri, Y.; Moreau, H.; Verger, R. Gastric lipases—Biochemical and physiological-studies. Biochim. Biophys. Acta 1989, 1006, 255–271. [Google Scholar] [CrossRef]
- Akoh, C.C.; Lee, G.C.; Liaw, Y.C.; Huang, T.H.; Shaw, J.F. GDSL family of serine esterases/lipases. Prog. Lipid Res. 2004, 43, 534–552. [Google Scholar] [CrossRef]
- Holm, C.; Kirchgessner, T.G.; Svenson, K.L.; Fredrikson, G.; Nilsson, S.; Miller, C.G.; Shively, J.E.; Heinzmann, C.; Sparkes, R.S.; Mohandas, T.; et al. Hormone-sensitive lipase—Sequence, expression, and chromosomal localization to 19 cent-q13.3. Science 1988, 241, 1503–1506. [Google Scholar] [CrossRef]
- Belfrage, P.; Fredrikson, G.; Stalfors, P.; Tornqvist, H. Lipases; Borgström, B., Brockman, H.I., Eds.; Elsevier: Amsterdam, The Netherlands, 1984; pp. 365–416. [Google Scholar]
- Hu, C. Life-history and occurrence of Pteromalus puparum L. in Hangzhou. Acta Entomol. Sin. 1984, 27, 302–307. [Google Scholar]
- Yan, Z.C.; Fang, Q.; Wang, L.; Liu, J.D.; Zhu, Y.; Wang, F.; Li, F.; Werren, J.H.; Ye, G.Y. Insights into the venom composition and evolution of an endoparasitoid wasp by combining proteomic and transcriptomic analyses. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.Y.; Fang, Q.; Wang, L.; Hu, C.; Ye, G.Y. Proteomic analysis of the venom from the endoparasitoid wasp Pteromalus puparum (Hymenoptera: Pteromalidae). Arch. Insect Biochem. Physiol. 2010, 75, 28–44. [Google Scholar] [CrossRef]
- Cusumano, A.; Duvic, B.; Jouan, V.; Ravallec, M.; Legeai, F.; Peri, E.; Colazza, S.; Volkoff, A.N. First extensive characterization of the venom gland from an egg parasitoid: Structure, transcriptome and functional role. J. Insect Physiol. 2018, 107, 68–80. [Google Scholar] [CrossRef]
- Laurino, S.; Grossi, G.; Pucci, P.; Flagiello, A.; Bufo, S.A.; Bianco, G.; Salvia, R.; Vinson, S.B.; Vogel, H.; Falabella, P. Identification of major Toxoneuron nigriceps venom proteins using an integrated transcriptomic/proteomic approach. Insect Biochem. Mol. Biol. 2016, 76, 49–61. [Google Scholar] [CrossRef]
- Mathe-Hubert, H.; Colinet, D.; Deleury, E.; Belghazi, M.; Ravallec, M.; Poulain, J.; Dossat, C.; Poirie, M.; Gatti, J.L. Comparative venomics of Psyttalia lounsburyi and P. concolor, two olive fruit fly parasitoids: A hypothetical role for a GH1 beta-glucosidase. Sci. Rep. 2016, 6, 35873. [Google Scholar] [CrossRef]
- De Graaf, D.C.; Aerts, M.; Brunain, M.; Desjardins, C.A.; Jacobs, F.J.; Werren, J.H.; Devreese, B. Insights into the venom composition of the ectoparasitoid wasp Nasonia vitripennis from bioinformatic and proteomic studies. Insect Mol. Biol. 2010, 19, 11–26. [Google Scholar] [CrossRef] [Green Version]
- Sim, A.D.; Wheeler, D. The venom gland transcriptome of the parasitoid wasp Nasonia vitripennis highlights the importance of novel genes in venom function. BMC Genom. 2016, 17, 571. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.Y.; Wang, J.Q.; Zhang, Z.B.; Huang, J.M.; Zhu, J.Y. Unraveling the venom components of an encyrtid endoparasitoid wasp Diversinervus elegans. Toxicon 2017, 136, 15–26. [Google Scholar] [CrossRef]
- Xin, B.; Liu, P.X.; Xu, X.R.; Zhang, S.; Zheng, Y.N. Identification of venom proteins of the indigenous endoparasitoid Chouioia cunea (Hymenoptera: Eulophidae). J. Econ. Entomol. 2017, 110, 2022–2030. [Google Scholar] [CrossRef]
- Vincent, B.; Kaeslin, M.; Roth, T.; Heller, M.; Poulain, J.; Cousserans, F.; Schaller, J.; Poirie, M.; Lanzrein, B.; Drezen, J.M.; et al. The venom composition of the parasitic wasp Chelonus inanitus resolved by combined expressed sequence tags analysis and proteomic approach. BMC Genom. 2010, 11, 693. [Google Scholar] [CrossRef] [Green Version]
- Heavner, M.E.; Gueguen, G.; Rajwani, R.; Pagan, P.E.; Small, C.; Govind, S. Partial venom gland transcriptome of a Drosophila parasitoid wasp, Leptopilina heterotoma, reveals novel and shared bioactive profiles with stinging Hymenoptera. Gene 2013, 526, 195–204. [Google Scholar] [CrossRef] [Green Version]
- Colinet, D.; Deleury, E.; Anselme, C.; Cazes, D.; Poulain, J.; Azema-Dossat, C.; Belghazi, M.; Gatti, J.L.; Poirie, M. Extensive inter- and intraspecific venom variation in closely related parasites targeting the same host: The case of Leptopilina parasitoids of Drosophila. Insect Biochem. Mol. Biol. 2013, 43, 601–611. [Google Scholar] [CrossRef]
- Burke, G.R.; Strand, M.R. Systematic analysis of a wasp parasitism arsenal. Mol. Ecol. 2014, 23, 890–901. [Google Scholar] [CrossRef]
- Akanbi, T.O.; Adcock, J.L.; Barrow, C.J. Selective concentration of EPA and DHA using Thermomyces lanuginosus lipase is due to fatty acid selectivity and not regioselectivity. Food Chem. 2013, 138, 615–620. [Google Scholar] [CrossRef]
- Haba, E.; Bresco, O.; Ferrer, C.; Marques, A.; Busquets, M.; Manresa, A. Isolation of lipase-secreting bacteria by deploying used frying oil as selective substrate. Enzym. Microb. Technol. 2000, 26, 40–44. [Google Scholar] [CrossRef]
- Hiol, A.; Jonzo, M.D.; Rugani, N.; Druet, D.; Sarda, L.; Comeau, L.C. Purification and characterization of an extracellular lipase from a thermophilic Rhizopus oryzae strain isolated from palm fruit. Enzym. Microb. Technol. 2000, 26, 421–430. [Google Scholar] [CrossRef]
- Olusesan, A.T.; Azura, L.K.; Forghani, B.; Abu Bakar, F.; Mohamed, A.K.S.; Radu, S.; Manap, M.Y.A.; Saari, N. Purification, characterization and thermal inactivation kinetics of a non-regioselective thermostable lipase from a genotypically identified extremophilic Bacillus subtilis NS 8. New Biotechnol. 2011, 28, 738–745. [Google Scholar] [CrossRef]
- Wang, J.; Schlenke, T.; Ye, G. Lipidomics reveals how the endoparasitoid wasp Pteromalus puparum manipulates host energy stores for its young. Entomology 2019, in press. [Google Scholar]
- Gao, X.K.; Zhang, S.; Luo, J.Y.; Lu, L.M.; Zhang, L.J.; Cui, J.J. Lipidomics and RNA-seq study of lipid regulation in Aphis gossypii parasitized by Lysiphlebia japonica. Sci. Rep. 2017, 7, 1–13. [Google Scholar]
- Kim, Y. Genome level analysis of Pteromalus puparum transcriptome: Preface. Arch. Insect Biochem. 2020, 103, e21641. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, 279–285. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Lin, Z.; Fang, Q.; Wang, J.L.; Yan, Z.C.; Zou, Z.; Song, Q.S.; Ye, G.Y. The genomic and transcriptomic analyses of serine proteases and their homologs in an endoparasitoid, Pteromalus puparum. Dev. Comp. Immunol. 2017, 77, 56–68. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT > method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Dodson, G.; Wlodawer, A. Catalytic triads and their relatives. Trends Biochem. Sci. 1998, 23, 347–352. [Google Scholar] [CrossRef]
- Horne, I.; Haritos, V.S.; Oakeshott, J.G. Comparative and functional genomics of lipases in holometabolous insects. Insect Biochem. Mol. Biol. 2009, 39, 547–567. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [Green Version]
- Abascal, F.; Zardoya, R.; Posada, D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 2005, 21, 2104–2105. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, 242–245. [Google Scholar] [CrossRef]
- Lowe, M.E. Properties and function of pancreatic lipase related protein 2. Biochimie 2000, 82, 997–1004. [Google Scholar] [CrossRef]
- Brenner, S. The molecular evolution of genes and proteins—A tale of 2 serines. Nature 1988, 334, 528–530. [Google Scholar] [CrossRef]
- Hide, W.A.; Chan, L.; Li, W.H. Structure and evolution of the lipase superfamily. J. Lipid Res. 1992, 33, 167–178. [Google Scholar]
- Paetzel, M.; Dalbey, R.E. Catalytic hydroxyl/amine dyads within serine proteases. Trends Biochem. Sci. 1997, 22, 28–31. [Google Scholar] [CrossRef]
- Valenzuela, J.G.; Francischetti, I.M.B.; Pham, V.M.; Garfield, M.K.; Ribeiro, J.M.C. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem. Mol. 2003, 33, 717–732. [Google Scholar] [CrossRef]
- Winkler, F.K.; Darcy, A.; Hunziker, W. Structure of human pancreatic lipase. Nature 1990, 343, 771–774. [Google Scholar] [CrossRef]
- Aoki, J.; Inoue, A.; Makide, K.; Saiki, N.; Arai, H. Structure and function of extracellular phospholipase A(1) belonging to the pancreatic lipase gene family. Biochimie 2007, 89, 197–204. [Google Scholar] [CrossRef]
- Hirata, K.; Dichek, H.L.; Cioffi, J.A.; Choi, S.Y.; Leeper, N.J.; Quintana, L.; Kronmal, G.S.; Cooper, A.D.; Quertermous, T. Cloning of a unique lipase from endothelial cells extends the lipase gene family. J. Biol. Chem. 1999, 274, 14170–14175. [Google Scholar] [CrossRef] [Green Version]
- Aoki, J.; Nagai, Y.; Hosono, H.; Inoue, K.; Arai, H. Structure and function of phosphatidylserine-specific phospholipase A1. Biochim. Biophys. Acta 2002, 1582, 26–32. [Google Scholar] [CrossRef]
- Hoffman, D.R.; Sakell, R.H.; Schmidt, M. Sol i 1, the phospholipase allergen of imported fire ant venom. J. Allergy Clin. Immunol. 2005, 115, 611–616. [Google Scholar] [CrossRef]
- Sukprasert, S.; Rungsa, P.; Uawonggul, N.; Incamnoi, P.; Thammasirirak, S.; Daduang, J.; Daduang, S. Purification and structural characterisation of phospholipase A(1) (Vespapase, Ves a 1) from Thai banded tiger wasp (Vespa affinis) venom. Toxicon 2013, 61, 151–164. [Google Scholar] [CrossRef]
- Perez-Riverol, A.; Pereira, F.D.C.; Lasa, A.M.; Fernandes, L.G.R.; dos Santos-Pinto, J.R.A.; Justo-Jacomini, D.L.; de Azevedo, G.O.; Bazon, M.L.; Palma, M.S.; Zollner, R.D.; et al. Molecular cloning, expression and IgE-immunoreactivity of phospholipase Al, a major allergen from Polybia paulista (Hymenoptera: Vespidae) venom. Toxicon 2016, 124, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Meadows, S.; Sharp, L.; Jan, L.Y.; Jan, Y.N. Genome-wide study of aging and oxidative stress response in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2000, 97, 13726–13731. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.; Aikins, J.; Wang, L.J.; Beeman, R.W.; Oppert, B.; Lord, J.C.; Brown, S.J.; Lorenzen, M.D.; Richards, S.; Weinstock, G.M.; et al. Analysis of transcriptome data in the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. 2008, 38, 380–386. [Google Scholar] [CrossRef] [Green Version]
- Dana, A.N.; Hillenmeyer, M.E.; Lobo, N.F.; Kern, M.K.; Romans, P.A.; Collins, F.H. Differential gene expression in abdomens of the malaria vector mosquito, Anopheles gambiae, after sugar feeding, blood feeding and Plasmodium berghei infection. BMC Genom. 2006, 7, 119. [Google Scholar] [CrossRef] [Green Version]
- Van Antwerpen, R.; Salvador, K.; Tolman, K.; Gentry, C. Uptake of lipids by developing oocytes of the hawkmoth Manduca sexta. The possible role of lipoprotein lipase. Insect Biochem. Mol. 1998, 28, 399–408. [Google Scholar] [CrossRef]
- Whitfield, C.W.; Band, M.R.; Bonaldo, M.F.; Kumar, C.G.; Liu, L.; Pardinas, J.R.; Robertson, H.M.; Soares, M.B.; Robinson, G.E. Annotated expressed sequence tags and cDNA microarrays for studies of brain and behavior in the honey bee. Genome Res. 2002, 12, 555–566. [Google Scholar] [CrossRef] [Green Version]
- Nardini, M.; Dijkstra, B.W. Alpha/beta hydrolase fold enzymes: The family keeps growing. Curr. Opin. Struct. Biol. 1999, 9, 732–737. [Google Scholar] [CrossRef]
- Ollis, D.L.; Cheah, E.; Cygler, M.; Dijkstra, B.; Frolow, F.; Franken, S.M.; Harel, M.; Remington, S.J.; Silman, I.; Schrag, J.; et al. The alpha/beta-hydrolase fold. Protein Eng. 1992, 5, 197–211. [Google Scholar] [CrossRef] [Green Version]
- Van Pouderoyen, G.; Eggert, T.; Jaeger, K.-E.; Dijkstra, B.W. The crystal structure of Bacillus subtili lipase: A minimal α/β hydrolase fold enzyme. J. Mol. Biol. 2001, 309, 215–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bownes, M. Why is there sequence similarity between insect yolk proteins and vertebrate lipases. J. Lipid Res. 1992, 33, 777–790. [Google Scholar] [PubMed]
- Hagedorn, H.H.; Maddison, D.R.; Tu, Z.J. The evolution of vitellogenins, cyclorrhaphan yolk proteins and related molecules. Adv. Insect Physiol. 1998, 27, 335–384. [Google Scholar]
- Amdam, G.V.; Norberg, K.; Hagen, A.; Omholt, S.W. Social exploitation of vitellogenin. Proc. Natl. Acad. Sci. USA 2003, 100, 1799–1802. [Google Scholar] [CrossRef] [Green Version]




| Species | Neutral | Acid | Lipase2 | Lipase3 | GDSL | Hormone Sensitive | Total |
|---|---|---|---|---|---|---|---|
| Pteromalus puparum | 32 | 26 | 0 | 3 | 2 | 1 | 64 |
| Nasonia vitripennis | 30 | 27 | 0 | 2 | 2 | 1 | 62 |
| Solenopsis invicta | 28 | 25 | 0 | 2 | 6 | 1 | 62 |
| Trichogramma pretiosum | 30 | 24 | 0 | 2 | 2 | 1 | 59 |
| Drosophila melanogaster1 | 31 | 21 | 0 | 1 | 2 | 1 | 56 |
| Tribolium castaneum1 | 25 | 25 | 0 | 1 | 2 | 1 | 54 |
| Anopheles gambiae1 | 28 | 14 | 0 | 1 | 7 | 1 | 51 |
| Diachasma alloeum | 18 | 22 | 0 | 3 | 4 | 1 | 48 |
| Copidosoma floridanum | 24 | 17 | 0 | 2 | 2 | 1 | 46 |
| Fopius arisanus | 14 | 19 | 0 | 2 | 2 | 1 | 38 |
| Ceratosolen solmsi | 16 | 11 | 0 | 2 | 1 | 1 | 31 |
| Bombyx mori1 | 11 | 14 | 0 | 1 | 2 | 1 | 29 |
| Orussus abietinus | 18 | 6 | 0 | 2 | 1 | 1 | 28 |
| Apis mellifera1 | 14 | 4 | 0 | 1 | 6 | 1 | 26 |
| Tissue | Gene ID | Position of Signal Peptide | PSMs | Tissue_FPKM | Log2(Vg_FPKM/Ca_FPKM) | Description | Accession |
|---|---|---|---|---|---|---|---|
| Venom gland | PPU04615 | 1–19 | N | 47.8084 | ∞ | PREDICTED: pancreatic lipase-related protein 2-like | XP_016836798.1 |
| PPU01585 | 1–16 | 7 | 107.410 | ∞ | PREDICTED: pancreatic lipase-related protein 2-like isoform X2 | XP_016845667.1 | |
| PPU09230 | N | 13 | 868.856 | 10.60339576 | PREDICTED: pancreatic lipase-related protein 2-like | XP_016838373.1 | |
| PPU11430 | 1–17 | 22 | 1041.80 | 10.1179628 | PREDICTED: gastric triacylglycerol lipase-like | XP_016840181.1 | |
| PPU16612 | 1–19 | 51 | 2022.12 | 9.693282524 | lipase A-like precursor | NP_001154991.1 | |
| PPU00494 | N | 5 | 1526.20 | 9.52790379 | PREDICTED: pancreatic lipase-related protein 2-like | XP_008215592.1 | |
| PPU04616 | 1–18 | 2 | 265.566 | 8.762219154 | PREDICTED: pancreatic lipase-related protein 2-like | XP_016836798.1 | |
| PPU09231 | 1–22 | 12 | 211.041 | 8.503220789 | PREDICTED: pancreatic lipase-related protein 2-like | XP_016838373.1 | |
| PPU10799 | N | N | 10.1819 | 0.929008269 | PREDICTED: pancreatic lipase-related protein 2-like | XP_008217694.1 | |
| PPU09657 | 1–21 | N | 37.3270 | 0.540844581 | PREDICTED: pancreatic lipase-related protein 2-like isoform X1 | XP_003425032.1 | |
| Larval salivary gland | PPU01336 | 1–20 | - | 113.132 | - | PREDICTED: lipase3-like | XP_016841509.1 |
| PPU10150 | 1–20 | - | 24.8997 | - | PREDICTED: lipase3-like | XP_008216710.1 | |
| PPU11431 | N | - | 5.76564 | - | PREDICTED: gastric triacylglycerol lipase-like | XP_016840181.1 | |
| PPU10742 | 1–20 | - | 0.517292 | - | PREDICTED: lipase3-like | XP_015112844.1 | |
| PPU13285 | 1–23 | - | 6.65252 | - | PREDICTED: pancreatic triacylglycerol lipase-like | XP_016844594.1 |
| Tracking_ID | Predicted Active Site Residues | β9 Loop | Length | Lid | Length |
|---|---|---|---|---|---|
| PPU01585 | S34D62- | - | 0 | - | 0 |
| PPU00494 | S16D44- | HTQGGKRDNNKAFGLNALLG | 20 | - | 0 |
| PPU16687 | S158D182H244 | HTNVDNCGMTYQVGH | 15 | CNSNRC | 6 |
| PPU16688 | S163D187H249 | HTDIQECGLKDQIGH | 15 | CEKHKC | 6 |
| PPU16689 | S191D215H280 | HTNAGLLGYLSAIGK | 15 | CSIDIGGSC | 9 |
| PPU13747 | S187D211H280 | HTCAGTVGFVRPIGH | 15 | CPVLMTQYC | 9 |
| PPU06394 | S164D188H254 | HTNGGNLGIRYPLGH | 15 | CGADLIGSC | 9 |
| PPU04615 | S177D206- | HTQTGNGEKISGFGLQKPSGH | 21 | CEIKSDGYV | 9 |
| PPU10689 | S171D195H265 | HTSGTAFGFLAAIGH | 15 | CNFAPTNTYC | 10 |
| PPU05374 | S170D194H259 | HTNAGYYGELGKVGH | 15 | CENRPNHELC | 10 |
| PPU06393 | S80D104H172 | HTNSNYYGLPEPRGH | 15 | CADEPNQKYYC | 11 |
| PPU10799 | S235D260H329 | HSCGGVLGFLQPLGH | 15 | CCCVPELIEAC | 11 |
| PPU05625 | S172D197H265 | HTGAGILGQWGPNGH | 15 | CATASLLQTLSC | 12 |
| PPU07948 | S164D188H258 | HTDGGIYGAYEPTGS | 15 | CFLFGVPLSPRGL | 13 |
| PPU09658 | S25D53I128 | HTQTGTGGSVDGLGLKESIGH | 21 | CVRKKVGDDYLKN | 13 |
| PPU06103 | S129D157Q232 | HTQTGNDEDISGIGVQERSGH | 21 | CETKHMTIQKMLC | 13 |
| PPU09657 | S176D204H279 | HTQTGTGGSVDGLGLKESIGH | 21 | CVSKTLKWDNMIC | 13 |
| PPU04616 | -D203H281 | HTQTGHGNGINAFGLEAPVGH | 21 | CEAKSIFYTEVNKMIC | 16 |
| PPU08363 | S174D203H283 | HTQTGNGNGINGLGLQESIGH | 21 | CERVSSIIHTTRIQKMIC | 18 |
| PPU01586 | - | HTDAVRTKNDEFGIRDPIGH | 20 | CDRRKRSFVTCWAMVAAIILEL | 22 |
| PPU13290 | S266D295H379 | HTNGQFLKKLGLGLPQPIGH | 20 | CELTSFTIPVLSIPREAINKAIC | 23 |
| PPU13286 | S111D144H229 | HTNAQNIMILGFGLPTQLGH | 20 | CAKIDTSFWDFLLLPVNIVKSAI | 23 |
| PPU13285 | S263D296H381 | HTNARQIYFLGLGLPEQLGF | 20 | CSDIDTSIWSFLLLPKTIIQESIC | 24 |
| PPU16276 | S193D220H308 | HTDGSVDFADGFGLLKPIGH | 20 | CKDVKNSVVVSHLNEDSLDIHIAC | 24 |
| PPU07195 | S208D237H322 | HTNAKGSLTEGLSLFKPIGH | 20 | CSESNFILPDSIKLPKRIIDEAVC | 24 |
| PPU09686 | S464D493H578 | HTNGRVLSKLGLGLPYPVGH | 20 | CILSETSLWRYLPLPIQKISETIC | 24 |
| PPU09687 | S229D258H343 | HTNGRILKKLGLGLPYPLGH | 20 | CILAKSSIWKYLPLPIEKIKKTIC | 24 |
| PPU06272 | S218D241H326 | HSNGEQLILGGLGSWQPMGD | 20 | CSNLFVGAVSDIIWSSPVEGRSLC | 24 |
| PPU03466 | S212D240H327 | HTDCSPFISGGLGISQPVAH | 20 | CNEGVFNSITLEKGSFFRGIKRFLGC | 26 |
| PPU03462 | S231D255H349 | HTDGKSIFFLGLPGYGMSQPCGH | 23 | CTDLSETTPSLPLTLIREGLEEASRVLVAC | 30 |
| PPU09230 | S71D97H202 | HTNSNPSGDTFGLYEPLGH | 19 | CSSNRVARTFSKDSVFIKCFSELFLGMDLSQLYGNIKSSRVELAK | 45 |
| PPU09231 | S71D98H208 | HTNSDPNRSTLGLYERLGH | 19 | CNNNQARTFSLWDAWEAIKNCTLIYFAGDNYSDLASEILTNLLTHSMVC | 49 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Song, J.; Fang, Q.; Yao, H.; Wang, F.; Song, Q.; Ye, G. Insight into the Functional Diversification of Lipases in the Endoparasitoid Pteromalus puparum (Hymenoptera: Pteromalidae) by Genome-scale Annotation and Expression Analysis. Insects 2020, 11, 227. https://doi.org/10.3390/insects11040227
Wang J, Song J, Fang Q, Yao H, Wang F, Song Q, Ye G. Insight into the Functional Diversification of Lipases in the Endoparasitoid Pteromalus puparum (Hymenoptera: Pteromalidae) by Genome-scale Annotation and Expression Analysis. Insects. 2020; 11(4):227. https://doi.org/10.3390/insects11040227
Chicago/Turabian StyleWang, Jiale, Jiqiang Song, Qi Fang, Hongwei Yao, Fang Wang, Qisheng Song, and Gongyin Ye. 2020. "Insight into the Functional Diversification of Lipases in the Endoparasitoid Pteromalus puparum (Hymenoptera: Pteromalidae) by Genome-scale Annotation and Expression Analysis" Insects 11, no. 4: 227. https://doi.org/10.3390/insects11040227