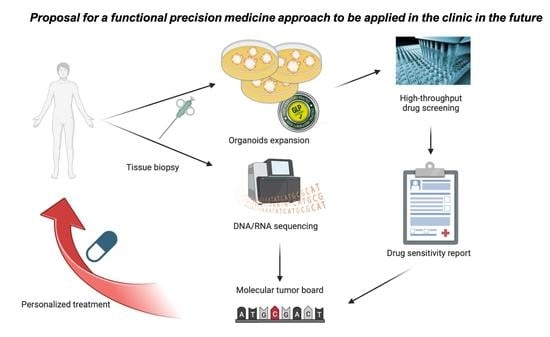
1. Introduction
In the last decades, huge efforts have been made on the molecular characterization of solid tumors, condensed in The Cancer Genome Atlas project. Since the introduction of the first molecularly targeted agent trastuzumab, an increasing number of drugs have been approved based on the presence of a particular molecular alteration, mostly represented by hotspot mutations or copy number variations in driver genes, opening the field for precision medicine in oncology. According to the European Society of Medical Oncology (ESMO), precision medicine can be defined as “a healthcare approach with the primary aim of identifying which interventions are likely to be of most benefit to which patients based upon the features of the individual and their disease. In cancer, the term usually refers to the use of therapeutics that are expected to confer benefit to a subset of patients whose cancer displays specific molecular or cellular features” [
1]; in summary “the right drug, to the right patient, at the right time”.
Recently, the introduction of patient-derived organoids (PDOs) (stable primary cultures derived directly from the patient) showed the possibility of using them to predict in vitro the response that should be observed in the patient. This could represent a way to test the effectiveness of a precision medicine approach before administering it to the patient, maximizing clinical benefit.
2. What Has the Clinical Impact of an Approach Based on Precision Medicine Been So Far?
Thanks to the implementation of modern next-generation sequencing (NGS) technologies and, in general, to improved knowledge of the molecular basis of neoplasms, over the past 20 years we have seen a disruptive and growing approval of molecularly targeted drugs. Alongside well-known molecular targets, new ones are constantly being identified, so much so that today there are more than 30 promising genomic targets. Such a list is just a first iteration of an ongoing process, and the number of targets is expected to continue to expand at a rapid pace. However, the final number of genes that could be drug targeted is difficult to predict. The cumulative work establishing these targets has already significantly increased the fraction of patients who are now believed to carry at least one potentially targetable genomic alteration. The number of tumors harboring at least one targetable alteration is constantly increasing, as is the number of potentially druggable alterations [
2], therefore increasing the complexity of molecularly guided treatment approach in solid tumors.
However, what is the clinical impact of an approach based on precision medicine? On closer inspection, the clinical studies conducted so far have proved to be rather disappointing, not being able to reach the set endpoints, either due to difficulties related to the often heavily pretreated study population, or to difficulties related to the execution times of genomic analyzes, etc. For instance, the phase 2 SHIVA trial [
3] randomized patients to receive a matched molecularly targeted agent or treatment according to physician’s choice. It did not show a benefit in terms of progression-free survival (PFS). Moreover, the MOSCATO-01 trial highlighted the fact that often a very small percentage of patients effectively benefit from this approach [
4]. Similar findings were also shown when liquid biopsy was used to detect molecular alterations in the TARGET study [
5]. Disappointing results in terms of objective response rate and feasibility were shown by the NCI-MPACT trial [
6].
Albeit substantially negative, from these studies a strong propulsive thrust emerges, evidenced by the exceptional responses seen in individual cases and based precisely on precision medicine approaches. Indeed, encouraging data come from the more recent MAST study [
7], where patients in the molecularly guided treatment arm had a higher PFS, and the percentage of patients able to receive further treatments was double in the experimental arm.
Collectively, the clinical impact of a precision medicine approach has not reached the level that was expected. One the most important issues is represented by the fact that most personalized oncology approaches rely on genomics, although it is of crucial relevance to consider that genomics does not automatically predict the response to a given treatment. Indeed, multiple factors can influence the response in the individual patient: the coexistence of other molecular alterations, gene expression, protein modifications, the tumor microenvironment, microbiota, the activation of alternative molecular pathways, and other less explored mechanisms. These factors contribute to interpatient heterogeneity [
7] in response, even when the same genomic alteration is shared.
This heterogeneity in response is certainly the epiphenomenon of the various mechanisms that contribute to the heterogeneity that is not only inter- but also intra-tumoral. Indeed, this year Hanahan added phenotypic plasticity, epigenetic reprogramming, senescence, and the role of the microbiota to the fundamental Hallmarks of Cancer work [
8].
3. The Need to Advance Functional Precision Medicine
In this context, precision medicine approaches should move from a static environment to a dynamic one, leading to the so-called functional precision medicine [
9]. This is focused on the possibility to treat patient-derived cancer cells with a given drug, which should be administered when an in vitro response is observed. This is why Letai proposes a new approach to precision medicine, which should take place through the integration of several techniques, where drugs are selected on the basis of genomic and functional tests performed directly on the tumor sample of the individual patient [
10]. Therefore, functional precision medicine can be defined as a strategy whereby live tumor cells of affected individuals are directly perturbed with drugs in order to provide personalized information that can be immediately translated to guide therapy [
9]. For this reason, it is essential to choose the most appropriate model to use.
Indeed, for many years this kind of approach has been technically impossible, as primary cell lines typically were cultured only for a few passages, therefore impeding drug screening approaches. Recently, this situation has been completely revolutionized by the development of a new primary culture technique, represented by the so-called organoids.
Organoids are 3D primary cultures that can be derived from both healthy and pathological patient tissue and can be grown over a long period of time, while maintaining genomic stability. They are in a more physiologically relevant condition than cell lines; cell-cell and cell-matrix interactions are preserved; they can be expanded, so they are perfectly suitable for drug screening; they can be used for genome editing; and they are perfect models for studying development and in particular human diseases. Therefore, they quickly became key models for translational research as well.
Organoids derived from human tissues have the advantage of being representative of human physiology, unlike animal models, which are only similar to it, they can be established in a quicker (compared to, for instance, patient-derived xenografts, PDXs) and simpler manner, they can be used for large-scale genetic screening as well as for drug screening, they can be genetically manipulated according to the most modern techniques, and since they are obtained by single individuals, they allow biological phenomena to be studied in a personalized way.
The development of organoids started thanks to identification of the Lgr5+ adult stem cells population within the intestinal crypt [
11], which led to the development of the growth factor recipe for human intestinal organoids [
12]. This opened the way to the generation of organoids from many tissues, through slight modification of organoid media composition, leading to the development of so-called “living biobanks” of organoids from many healthy and diseased tissues.
Human organoids are increasing knowledge in the field of oncology, thanks to the isolation of organoids derived from tumor tissues. Recent studies describe the genomic and morphological characterization of tumor organoids from biopsies of colon [
13,
14,
15,
16,
17], brain [
18], prostate [
19], pancreas [
20,
21], liver [
22], breast [
23], stomach [
24], esophagus [
25], endometrium [
26], lung [
27,
28], and the head and neck [
29]. These studies have shown that organoids maintain the same characteristics of the original tissue in culture; for instance, showing that PDOs maintain the same immunohistochemical markers of original tissue, therefore showing that the cell population is similar. In particular, while all the studies reported on genomic profile, showing that PDOs replicate both the mutation and copy number profile, only a few studies have reported on the transcriptomic similarities [
25] and none of them have reported on proteomics. More relevant for precision medicine, many studies showed that PDOs can replicate the response to pharmacological treatments, with the study by Vlachogiannis et al. reporting an outstanding 100% sensitivity, 93% specificity, 88% positive predictive value, and 100% negative predictive value for PDOs in predicting response in patients [
17].
4. The Impact of Organoids for Functional Precision Medicine
The implementation of PDOs could represent the key towards functional precision medicine, allowing in the future non-responding patients to be spared side effects in the case of drugs that are already in the clinic or at an advanced development stage, but also allowing for the functional validation of emerging genomic and non-genomic biomarkers and the potential to screen new active compounds in models that are a faithful reproduction of the tumor, therefore potentially shortening the path of drug development.
Clearly, this technology is in its infancy, although it is extremely promising. The development of organoids from adult tissues is limited by the accessibility of the tissue of interest and by the knowledge of the culture conditions of that particular tissue. Nevertheless, there are some disadvantages, which can be interpreted as challenges for improving the technology: the absence of the microenvironment, particularly in organoids derived from adult stem cells, and the absence of standardized protocols and quality controls for which results may vary between research groups. Although cheaper than mouse models, they are nevertheless much more expensive than cell lines. Furthermore, studies at the organ level are complex.
Perhaps what is most important for the implementation of organoids as a functional precision medicine tool, more than a “simple” genomic similarity, is the proof that they “behave” just like the patient’s tumor, specifically their capacity to replicate and predict treatment outcome. A good starting point and example is represented by what has been done for colorectal cancer (CRC). In a 2019 paper, a library of PDOs derived from metastatic CRC patients has been shown to predict response to irinotecan-based chemotherapy and not to 5-fluorouracil plus oxaliplatin [
30]. Therefore, these authors suggested that PDOs should be used to select those patients who could benefit from irinotecan from those who would only experience toxicity. In a more recent work, authors aimed at studying heterogeneity in the metastatic setting from a functional point of view in order to test if different PDOs lines derived from different metastatic sites of the same patient would respond differently to drug treatment. This is one of the key clinical questions, as mixed responses are often observed in the clinic. Surprisingly, intra-patient inter-metastatic heterogeneity in drug response was not observed [
16].
However, all these studies (and those conducted for other tumor types) were observational. The first interventional clinical study was the single-center, single-arm SENSOR study [
31] with the aim of evaluating the feasibility of PDOs to assign patients to treatment with off-label or experimental agents. The primary endpoint was an objective response rate greater than or equal to 20%. Patients underwent a culture biopsy before starting their last standard of care course. The organoids were exposed to a panel of eight drugs targeting main driver pathways, and patients were treated after progression on standard treatment after having identified a clear signal of antitumor activity in vitro. Sixty-one patients were included, and thirty-one organoids were generated from fifty-four eligible patients. Twenty-five cultures were screened for drugs and nineteen organoids showed substantial responses to one or more drugs. Three patients were treated with the mTOR inhibitor vistusertib and three were treated with the pan-Akt inhibitor capivasertib. Despite the drug sensitivity shown by organoids, patients did not demonstrate objective clinical responses to the recommended treatment. The authors concluded that this strategy is not feasible. However, of the 19 patients for whom a significant activity was shown for at least one drug in the PDO, only six started a treatment, while most of the remaining ones dropped out of the study due to clinical deterioration. Maybe the disease setting (patients without curative treatment options, heavily pretreated) is something that per se limited the accessibility of the treatment, together with the 10 weeks necessary to perform all the procedures, from PDOs generation to drug screening. What we can learn from this experience is the necessity to invest in improving the method: for instance, accelerating the process of developing organoids and maximize the performance of drug screening.
Another relevant setting for which organoids could give a significant contribution is represented by locally advanced rectal cancer (LARC), where treatment choice relies on clinical and radiological features [
32,
33] without biomarkers of response available, and where organ-preserving strategies are under closer evaluation. In this context, the possibility to predict patient response could be crucial to spare unnecessary side effects, plan a better treatment strategy, and to better select patients for conservative strategies.
For instance, Ganesh et al. [
34] established a biobank of pre- and post-treatment rectal cancer organoids with a success rate of 77%. Forty-nine lines were generated using tissue obtained with biopsy routinely used in clinical care. Of these, 22 were derived from treatment-naïve patients. Organoids were treated with both standard chemotherapy and radiation therapy, with a good correlation with patient’s response.
In a following larger study by Yao et al. [
35], organoids were generated from LARC patients before any treatment, establishing a total of 92 lines from 112 patients. Eighty lines were treated with radiation, 5-fluorouracil, and irinotecan. Interestingly, PDOs were able to reproduce clinical response, and patients achieved a good response when their organoids were sensitive to at least one agent. These data are encouraging, although no clinical data exist for single agent use nor for 5-fluorouracil and irinotecan combinations.
Clearly, future studies should better replicate the treatment strategies that are part of the standard treatment for LARC.
5. Future Perspectives
Of importance, there is a great need to standardize the procedures to develop and maintain organoids as well as to subject them to drug screening, otherwise results will never be comparable. To achieve this, a great effort is needed from both academic and pharma industries, which can be attracted by the great potential of this platform even for drug development. The value of PDOs for drug development has been recently proven in a paradigmatic study where CRC PDOs were employed for large-scale screening compounds of bispecific antibodies targeting Wnt and RTK and resulting in the development of an effective compound, MCLA-158, capable of inducing EGFR degradation in Lgr5+ cancer stem cells and inhibiting the growth of KRAS mutant CRC [
36]. These unprecedented results should encourage others to apply this approach to drug development in a systematic way.
Nevertheless, a lot of questions remain unanswered. First, uniform protocols are needed to homogenize and standardize culture conditions and success rates, which are quite different among different research groups, and to select the best conditions for drug screening and drug screening interpretation. A key issue is represented by the way to correlate in vitro with in vivo response. Current models are deprived of stromal and immune microenvironment; therefore, its possible effect on drug response is lost. Many efforts are undergoing to develop co-cultures that will allow, for instance, the incorporation of immunotherapy agents into drug screening platforms.
Clearly, the technology is still in its infancy, and many additional studies are needed to develop its use in functional precision medicine. The clinical trials of the future could include, in addition to an extensive molecular characterization, the analysis of PDO response to guide the allocation of patients to different treatment arms.
Author Contributions
Conceptualization, F.P., J.C., A.C.; methodology, F.P., J.C.; resources, M.C.-S., B.G.-M., N.T., D.R.; data curation, F.P., M.C.-S.; writing-original draft preparation, F.P.; writing-review and editing, F.P., M.C.-S., B.G.-M., N.T., D.R., J.C., A.C.; supervision, J.C., A.C.; funding acquisition, N.T., D.R., A.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Carlos III Health Institute [grant number PI18/01909 and PI21/00689] to A.C. and N.T. N.T. was funded by Joan Rodés contract JR20/00005 from the Carlos III Health Institute. D.R. was funded by Joan Rodés contract JR16/00040 from the Carlos III Health Institute. J.C. was awarded a VLC Bioclinic grant 2021/257 from the University of Valencia. M.C.-S. is supported by a grant from AECC (PRDVA19006CABE). F.P. was supported by ESMO Translational Research Project with the aid of a grant from Amgen; any views, opinions, findings, conclusions, or recommendations expressed in this material are those solely of the authors and do not necessarily reflect those of ESMO or Amgen.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
A.C. declares institutional research funding from Genentech, Merck Serono, BMS, MSD, Roche, Beigene, Bayer, Servier, Eli-Lilly, Novartis, Takeda, Astellas, Natera, and Fibrogen and advisory board or speaker fees from Amgen, Merck Serono, Roche, Bayer, Servier, and Pierre Fabre in the last five years. D.R. declares institutional research funding from Tesaro, AbbVie, Novartis, Merus, Roche, Relay Therapeutics, and HiFiBiO inc. F.P. declares institutional research funding from Merck Serono. All remaining authors have declared no conflict of interest. All the funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Yates, L.R.; Seoane, J.; Le Tourneau, C.; Siu, L.L.; Marais, R.; Michiels, S.; Soria, J.C.; Campbell, P.; Normanno, N.; Scarpa, A.; et al. The European Society for Medical Oncology (ESMO) Precision Medicine Glossary. Ann. Oncol. 2018, 29, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Hyman, D.M.; Taylor, B.S.; Baselga, J. Implementing Genome-Driven Oncology. Cell 2017, 168, 584–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Tourneau, C.; Delord, J.-P.; Gonçalves, A.; Gavoille, C.; Dubot, C.; Isambert, N.; Campone, M.; Trédan, O.; Massiani, M.-A.; Mauborgne, C.; et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): A multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015, 16, 1324–1334. [Google Scholar] [CrossRef]
- Massard, C.; Michiels, S.; Ferté, C.; Le Deley, M.-C.; Lacroix, L.; Hollebecque, A.; Verlingue, L.; Ileana, E.; Rosellini, S.; Ammari, S.; et al. High-Throughput Genomics and Clinical Outcome in Hard-to-Treat Advanced Cancers: Results of the MOSCATO 01 Trial. Cancer Discov. 2017, 7, 586–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothwell, D.G.; Ayub, M.; Cook, N.; Thistlethwaite, F.; Carter, L.; Dean, E.; Smith, N.; Villa, S.; Dransfield, J.; Clipson, A.; et al. Utility of ctDNA to support patient selection for early phase clinical trials: The TARGET study. Nat. Med. 2019, 25, 738–743. [Google Scholar] [CrossRef] [Green Version]
- Chen, A.P.; Kummar, S.; Moore, N.; Rubinstein, L.V.; Zhao, Y.; Williams, P.M.; Palmisano, A.; Sims, D.; O’Sullivan Coyne, G.; Rosenberger, C.L.; et al. Molecular Profiling-Based Assignment of Cancer Therapy (NCI-MPACT): A Randomized Multicenter Phase II Trial. JCO Precis. Oncol. 2021, 5, 133–144. [Google Scholar] [CrossRef]
- Gambardella, V.; Lombardi, P.; Carbonell-Asins, J.A.; Tarazona, N.; Cejalvo, J.M.; González-Barrallo, I.; Martín-Arana, J.; Tébar-Martínez, R.; Viala, A.; Bruixola, G.; et al. Molecular profiling of advanced solid tumours. The impact of experimental molecular-matched therapies on cancer patient outcomes in early-phase trials: The MAST study. Br. J. Cancer 2021, 125, 1261–1269. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Letai, A.; Bhola, P.; Welm, A.L. Functional precision oncology: Testing tumors with drugs to identify vulnerabilities and novel combinations. Cancer Cell 2022, 40, 26–35. [Google Scholar] [CrossRef]
- Letai, A. Functional precision cancer medicine—Moving beyond pure genomics. Nat. Med. 2017, 23, 1028–1035. [Google Scholar] [CrossRef]
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.J.; Van Es, J.H.; Van Den Brink, S.; Van Houdt, W.J.; Pronk, A.; Van Gorp, J.; Siersema, P.D.; et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef]
- Van De Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; Van Houdt, W.; Van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef] [Green Version]
- Buzzelli, J.N.; Ouaret, D.; Brown, G.; Allen, P.D.; Muschel, R.J. Colorectal cancer liver metastases organoids retain characteristics of original tumor and acquire chemotherapy resistance. Stem Cell Res. 2018, 27, 109–120. [Google Scholar] [CrossRef]
- Weeber, F.; Van De Wetering, M.; Hoogstraat, M.; Dijkstra, K.K.; Krijgsman, O.; Kuilman, T.; Gadellaa-Van Hooijdonk, C.G.M.; Van Der Velden, D.L.; Peeper, D.S.; Cuppen, E.P.J.G.; et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc. Natl. Acad. Sci. USA 2015, 112, 13308–13311. [Google Scholar] [CrossRef] [Green Version]
- Bruun, J.; Kryeziu, K.; Eide, P.W.; Moosavi, S.H.; Eilertsen, I.A.; Langerud, J.; Røsok, B.; Totland, M.Z.; Brunsell, T.H.; Pellinen, T.; et al. Patient-Derived Organoids from Multiple Colorectal Cancer Liver Metastases Reveal Moderate Intra-patient Pharmacotranscriptomic Heterogeneity. Clin. Cancer Res. 2020, 26, 4107–4119. [Google Scholar] [CrossRef] [Green Version]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernández-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef] [Green Version]
- Jacob, F.; Salinas, R.D.; Zhang, D.Y.; Nguyen, P.T.T.; Schnoll, J.G.; Wong, S.Z.H.; Thokala, R.; Sheikh, S.; Saxena, D.; Prokop, S.; et al. A Patient-Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter- and Intra-tumoral Heterogeneity. Cell 2020, 180, 188–204.e22. [Google Scholar] [CrossRef]
- Gao, D.; Vela, I.; Sboner, A.; Iaquinta, P.J.; Karthaus, W.R.; Gopalan, A.; Dowling, C.; Wanjala, J.N.; Undvall, E.A.; Arora, V.K.; et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014, 159, 176–187. [Google Scholar] [CrossRef] [Green Version]
- Driehuis, E.; van Hoeck, A.; Moore, K.; Kolders, S.; Francies, H.E.; Gulersonmez, M.C.; Stigter, E.C.A.; Burgering, B.; Geurts, V.; Gracanin, A.; et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc. Natl. Acad. Sci. USA 2019, 116, 26580–26590. [Google Scholar] [CrossRef]
- Seino, T.; Kawasaki, S.; Shimokawa, M.; Tamagawa, H.; Toshimitsu, K.; Fujii, M.; Ohta, Y.; Matano, M.; Nanki, K.; Kawasaki, K.; et al. Human Pancreatic Tumor Organoids Reveal Loss of Stem Cell Niche Factor Dependence during Disease Progression. Cell Stem Cell 2018, 22, 454–467.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broutier, L.; Mastrogiovanni, G.; Verstegen, M.M.; Francies, H.E.; Gavarró, L.M.; Bradshaw, C.R.; Allen, G.E.; Arnes-Benito, R.; Sidorova, O.; Gaspersz, M.P.; et al. Human primary liver cancer–derived organoid cultures for disease modeling and drug screening. Nat. Med. 2017, 23, 1424–1435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sachs, N.; de Ligt, J.; Kopper, O.; Gogola, E.; Bounova, G.; Weeber, F.; Balgobind, A.V.; Wind, K.; Gracanin, A.; Begthel, H.; et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell 2018, 172, 373–386.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, H.H.N.; Siu, H.C.; Law, S.; Ho, S.L.; Yue, S.S.K.; Tsui, W.Y.; Chan, D.; Chan, A.S.; Ma, S.; Lam, K.O.; et al. A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening. Cell Stem Cell 2018, 23, 882–897.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Francies, H.E.; Secrier, M.; Perner, J.; Miremadi, A.; Galeano-Dalmau, N.; Barendt, W.J.; Letchford, L.; Leyden, G.M.; Goffin, E.K.; et al. Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics. Nat. Commun. 2018, 9, 2983. [Google Scholar] [CrossRef] [Green Version]
- Boretto, M.; Maenhoudt, N.; Luo, X.; Hennes, A.; Boeckx, B.; Bui, B.; Heremans, R.; Perneel, L.; Kobayashi, H.; Van Zundert, I.; et al. Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat. Cell Biol. 2019, 21, 1041–1051. [Google Scholar] [CrossRef]
- Shi, R.; Radulovich, N.; Ng, C.; Liu, N.; Notsuda, H.; Cabanero, M.; Martins-Filho, S.N.; Raghavan, V.; Li, Q.; Mer, A.S.; et al. Organoid Cultures as Preclinical Models of Non–Small Cell Lung Cancer. Clin. Cancer Res. 2020, 26, 1162–1174. [Google Scholar] [CrossRef] [Green Version]
- Di Liello, R.; Ciaramella, V.; Barra, G.; Venditti, M.; Della Corte, C.M.; Papaccio, F.; Sparano, F.; Viscardi, G.; Iacovino, M.L.; Minucci, S.; et al. Ex vivo lung cancer spheroids resemble treatment response of a patient with NSCLC to chemotherapy and immunotherapy: Case report and translational study. ESMO Open 2019, 4, e000536. [Google Scholar] [CrossRef] [Green Version]
- Driehuis, E.; Kolders, S.; Spelier, S.; Lõhmussaar, K.; Willems, S.M.; Devriese, L.A.; de Bree, R.; de Ruiter, E.J.; Korving, J.; Begthel, H.; et al. Oral Mucosal Organoids as a Potential Platform for Personalized Cancer Therapy. Cancer Discov. 2019, 9, 852–871. [Google Scholar] [CrossRef]
- Ooft, S.N.; Weeber, F.; Dijkstra, K.K.; McLean, C.M.; Kaing, S.; van Werkhoven, E.; Schipper, L.; Hoes, L.; Vis, D.J.; van de Haar, J.; et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 2019, 11, eaay2574. [Google Scholar] [CrossRef]
- Ooft, S.N.; Weeber, F.; Schipper, L.; Dijkstra, K.K.; McLean, C.M.; Kaing, S.; van de Haar, J.; Prevoo, W.; van Werkhoven, E.; Snaebjornsson, P.; et al. Prospective experimental treatment of colorectal cancer patients based on organoid drug responses. ESMO Open 2021, 6, 100103. [Google Scholar] [CrossRef]
- Roselló, S.; Papaccio, F.; Roda, D.; Tarazona, N.; Cervantes, A. The role of chemotherapy in localized and locally advanced rectal cancer: A systematic revision. Cancer Treat. Rev. 2018, 63, 156–171. [Google Scholar] [CrossRef]
- Papaccio, F.; Roselló, S.; Huerta, M.; Gambardella, V.; Tarazona, N.; Fleitas, T.; Roda, D.; Cervantes, A. Neoadjuvant Chemotherapy in Locally Advanced Rectal Cancer. Cancers 2020, 12, 3611. [Google Scholar] [CrossRef]
- Ganesh, K.; Wu, C.; O’Rourke, K.P.; Szeglin, B.C.; Zheng, Y.; Sauvé, C.-E.G.; Adileh, M.; Wasserman, I.; Marco, M.R.; Kim, A.S.; et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat. Med. 2019, 25, 1607–1614. [Google Scholar] [CrossRef]
- Yao, Y.; Xu, X.; Yang, L.; Zhu, J.; Wan, J.; Shen, L.; Xia, F.; Fu, G.; Deng, Y.; Pan, M.; et al. Patient-Derived Organoids Predict Chemoradiation Responses of Locally Advanced Rectal Cancer. Cell Stem Cell 2020, 26, 17–26.e6. [Google Scholar] [CrossRef]
- Herpers, B.; Eppink, B.; James, M.I.; Cortina, C.; Cañellas-Socias, A.; Boj, S.F.; Hernando-Momblona, X.; Glodzik, D.; Roovers, R.C.; van de Wetering, M.; et al. Functional patient-derived organoid screenings identify MCLA-158 as a therapeutic EGFR × LGR5 bispecific antibody with efficacy in epithelial tumors. Nat. Cancer 2022, 3, 418–436. [Google Scholar] [CrossRef]
| Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).