Potential Role of miRNAs in the Acquisition of Chemoresistance in Neuroblastoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Cultures
2.2. Patient Samples
2.3. RNA Extraction
2.4. MiRNA Microarray Analysis
2.5. Real Time PCR Analysis
2.6. Comparative Genomic Hybridization (CGH) Analysis
2.7. PCA Analysis
2.8. Statistical Analysis
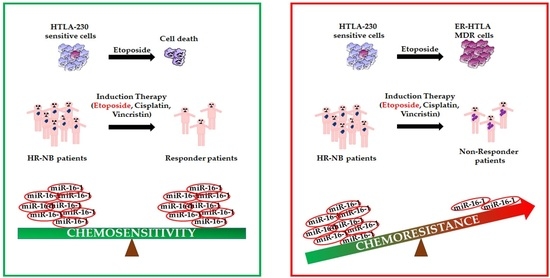
3. Results and Discussion
3.1. miRNA Expression Profiling of HTLA-230 and ER-HTLA Cells.
3.2. Comparative Genomic Hybridization (CGH) on HTLA-230 and ER-HTLA Cells
3.3. miRNA Expression Profiling of Therapy-Sensitive (Responder) and Therapy-Resistant (Non-Responder) NB Patients
3.4. miRNA Expression Profiling of NB Primary Tumors and Metastases
3.5. Principal Component Analysis (PCA) of the Results Obtained in Patients’ Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maris, J.M. Recent advances in neuroblastoma. N. Engl. J. Med. 2010, 362, 2202–2211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brodeur, G.M. Neuroblastoma: Biological insights into a clinical enigma. Nat. Rev. Cancer 2003, 3, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Pinto, N.R.; Applebaum, M.A.; Volchenboum, S.L.; Matthay, K.K.; London, W.B.; Ambros, P.F.; Nakagawara, A.; Berthold, F.; Schleiermacher, G.; Park, J.R.; et al. Advances in Risk Classification and Treatment Strategies for Neuroblastoma. J. Clin. Oncol. 2015, 33, 3008–3017. [Google Scholar] [CrossRef]
- Hochheuser, C.; van Zogchel, L.M.J.; Kleijer, M.; Kuijk, C.; Tol, S.; van der Schoot, C.E.; Voermans, C.; Tytgat, G.A.M.; Timmerman, I. The Metastatic Bone Marrow Niche in Neuroblastoma: Altered Phenotype and Function of Mesenchymal Stromal Cells. Cancers 2020, 12, 3231. [Google Scholar] [CrossRef]
- Seeger, R.C.; Reynolds, C.P.; Gallego, R.; Stram, D.O.; Gerbing, R.B.; Matthay, K.K. Quantitative tumor cell content of bone marrow and blood as a predictor of outcome in stage IV neuroblastoma: A Children’s Cancer Group Study. J. Clin. Oncol. 2000, 18, 4067–4076. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.Y.; Pan, C.; Tang, Y.J.; Chen, J.; Ye, Q.D.; Zhou, M.; Xue, H.; Tang, J.Y. Minimal residual disease is a prognostic marker for neuroblastoma with bone marrow infiltration. Am. J. Clin. Oncol. 2012, 35, 275–278. [Google Scholar] [CrossRef]
- Stallings, R.L. Are chromosomal imbalances important in cancer? Trends Genet. 2007, 23, 278–283. [Google Scholar] [CrossRef]
- Brodeur, G.M.; Seeger, R.C.; Schwab, M.; Varmus, H.E.; Bishop, J.M. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 1984, 224, 1121–1124. [Google Scholar] [CrossRef]
- Lin, R.J.; Lin, Y.C.; Chen, J.; Kuo, H.H.; Chen, Y.Y.; Diccianni, M.B.; London, W.B.; Chang, C.H.; Yu, A.L. microRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res. 2010, 70, 7841–7850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulte, J.H.; Schowe, B.; Mestdagh, P.; Kaderali, L.; Kalaghatgi, P.; Schlierf, S.; Vermeulen, J.; Brockmeyer, B.; Pajtler, K.; Thor, T.; et al. Accurate prediction of neuroblastoma outcome based on miRNA expression profiles. Int. J. Cancer 2010, 127, 2374–2385. [Google Scholar] [CrossRef]
- Mohammadi, M.; Goodarzi, M.; Jaafari, M.R.; Mirzaei, H.R.; Mirzaei, H. Circulating microRNA: A new candidate for diagnostic biomarker in neuroblastoma. Cancer Gene Ther. 2016, 23, 371–372. [Google Scholar] [CrossRef]
- Colla, R.; Izzotti, A.; De Ciucis, C.; Fenoglio, D.; Ravera, S.; Speciale, A.; Ricciarelli, R.; Furfaro, A.L.; Pulliero, A.; Passalacqua, M.; et al. Glutathione-mediated antioxidant response and aerobic metabolism: Two crucial factors involved in determining the multi-drug resistance of high-risk neuroblastoma. Oncotarget 2016, 7, 70715–70737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marengo, B.; Monti, P.; Miele, M.; Menichini, P.; Ottaggio, L.; Foggetti, G.; Pulliero, A.; Izzotti, A.; Speciale, A.; Garbarino, O.; et al. Etoposide-resistance in a neuroblastoma model cell line is associated with 13q14.3 mono-allelic deletion and miRNA-15a/16-1 down-regulation. Sci. Rep. 2018, 8, 13762. [Google Scholar] [CrossRef] [PubMed]
- Scaruffi, P.; Morandi, F.; Gallo, F.; Stigliani, S.; Parodi, S.; Moretti, S.; Bonassi, S.; Fardin, P.; Garaventa, A.; Zanazzo, G.; et al. Bone marrow of neuroblastoma patients shows downregulation of CXCL12 expression and presence of IFN signature. Pediatr. Blood Cancer 2012, 59, 44–51. [Google Scholar] [CrossRef]
- Morandi, F.; Scaruffi, P.; Gallo, F.; Stigliani, S.; Moretti, S.; Bonassi, S.; Gambini, C.; Mazzocco, K.; Fardin, P.; Haupt, R.; et al. Bone marrow-infiltrating human neuroblastoma cells express high levels of calprotectin and HLA-G proteins. PLoS ONE 2012, 7, e29922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stracquadanio, M.; Dinelli, E.; Trombini, C. Role of volcanic dust in the atmospheric transport and deposition of polycyclic aromatic hydrocarbons and mercury. J. Environ. Monit. 2003, 5, 984–988. [Google Scholar] [CrossRef]
- Alexandrov, K.; Rojas, M.; Geneste, O.; Castegnaro, M.; Camus, A.M.; Petruzzelli, S.; Giuntini, C.; Bartsch, H. An improved fluorometric assay for dosimetry of benzo(a)pyrene diol-epoxide-DNA adducts in smokers’ lung: Comparisons with total bulky adducts and aryl hydrocarbon hydroxylase activity. Cancer Res. 1992, 52, 6248–6253. [Google Scholar]
- Torres, A.; Torres, K.; Wdowiak, P.; Paszkowski, T.; Maciejewski, R. Selection and validation of endogenous controls for microRNA expression studies in endometrioid endometrial cancer tissues. Gynecol. Oncol. 2013, 130, 588–594. [Google Scholar] [CrossRef] [Green Version]
- Davies, T.; Fearn, T. Back to Basics: The Principles of Principal Component Analysis; Spectroscopy Europe: Charlston, Chichester, UK, 2004; p. 20. [Google Scholar]
- Leardi, R. Chemometric methods in food authentication. In Modern Techniques for Food Authentication, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 687–729. [Google Scholar]
- Leardi, R.; Melzi, C.; Polotti, G. CAT (Chemometric Agile Software). Available online: http://gruppochemiometria.it/index.php/software (accessed on 18 December 2020).
- Chava, S.; Reynolds, C.P.; Pathania, A.S.; Gorantla, S.; Poluektova, L.Y.; Coulter, D.W.; Gupta, S.C.; Pandey, M.K.; Challagundla, K.B. miR-15a-5p, miR-15b-5p, and miR-16-5p inhibit tumor progression by directly targeting MYCN in neuroblastoma. Mol. Oncol. 2020, 14, 180–196. [Google Scholar] [CrossRef]
- Pouliot, L.M.; Chen, Y.C.; Bai, J.; Guha, R.; Martin, S.E.; Gottesman, M.M.; Hall, M.D. Cisplatin sensitivity mediated by WEE1 and CHK1 is mediated by miR-155 and the miR-15 family. Cancer Res. 2012, 72, 5945–5955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soriano, A.; París-Coderch, L.; Jubierre, L.; Martínez, A.; Zhou, X.; Piskareva, O.; Bray, I.; Vidal, I.; Almazán-Moga, A.; Molist, C.; et al. MicroRNA-497 impairs the growth of chemoresistant neuroblastoma cells by targeting cell cycle, survival and vascular permeability genes. Oncotarget 2016, 7, 9271–9287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, C.; Gong, M.; Li, Z. Knockdown of lncRNA MCM3AP-AS1 Attenuates Chemoresistance of Burkitt Lymphoma to Doxorubicin Treatment via Targeting the miR-15a/EIF4E Axis. Cancer Manag. Res. 2020, 12, 5845–5855. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Fesler, A.; Huang, W.; Wang, Y.; Yang, J.; Wang, X.; Zheng, Y.; Hwang, G.R.; Wang, H.; Ju, J. Functional Significance and Therapeutic Potential of miR-15a Mimic in Pancreatic Ductal Adenocarcinoma. Mol. Ther. Nucleic Acids 2020, 19, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Fesler, A.; Liu, H.; Ju, J. Modified miR-15a has therapeutic potential for improving treatment of advanced stage colorectal cancer through inhibition of BCL2, BMI1, YAP1 and DCLK1. Oncotarget 2017, 9, 2367–2383. [Google Scholar] [CrossRef] [Green Version]
- Dwivedi, S.K.; Mustafi, S.B.; Mangala, L.S.; Jiang, D.; Pradeep, S.; Rodriguez-Aguayo, C.; Ling, H.; Ivan, C.; Mukherjee, P.; Calin, G.A.; et al. Therapeutic evaluation of microRNA-15a and microRNA-16 in ovarian cancer. Oncotarget 2016, 7, 15093–15104. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, J.; Liu, Q.; Zhao, Y.; Zhang, W.; Yang, H. Circ-CUX1 Accelerates the Progression of Neuroblastoma via miR-16-5p/DMRT2 Axis. Neurochem. Res. 2020, 45, 2840–2855. [Google Scholar] [CrossRef]
- Zhao, Z.; Ji, M.; Wang, Q.; He, N.; Li, Y. miR-16-5p/PDK4-Mediated Metabolic Reprogramming Is Involved in Chemoresistance of Cervical Cancer. Mol. Ther. Oncolytics 2020, 17, 509–517. [Google Scholar] [CrossRef]
- Patel, N.; Garikapati, K.R.; Pandita, R.K.; Singh, D.K.; Pandita, T.K.; Bhadra, U.; Bhadra, M.P. miR-15a/miR-16 down-regulates BMI1, impacting Ub-H2A mediated DNA repair and breast cancer cell sensitivity to doxorubicin. Sci. Rep. 2017, 7, 4263. [Google Scholar] [CrossRef] [PubMed]
- Venturutti, L.; Russo, R.I.C.; Rivas, M.A.; Mercogliano, M.F.; Izzo, F.; Oakley, R.H.; Pereyra, M.G.; De Martino, M.; Proietti, C.J.; Yankilevich, P.; et al. MiR-16 mediates trastuzumab and lapatinib response in ErbB-2-positive breast and gastric cancer via its novel targets CCNJ and FUBP1. Oncogene 2016, 35, 6189–6202. [Google Scholar] [CrossRef]
- Xia, L.; Zhang, D.; Du, R.; Pan, Y.; Zhao, L.; Sun, S.; Hong, L.; Liu, J.; Fan, D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int. J. Cancer 2008, 123, 372–379. [Google Scholar] [CrossRef]
- Chatterjee, A.; Chattopadhyay, D.; Chakrabarti, G. MiR-16 targets Bcl-2 in paclitaxel-resistant lung cancer cells and overexpression of miR-16 along with miR-17 causes unprecedented sensitivity by simultaneously modulating autophagy and apoptosis. Cell. Signal. 2015, 27, 189–203. [Google Scholar] [CrossRef]
- Liu, Y.; Gu, S.; Li, H.; Wang, J.; Wei, C.; Liu, Q. SNHG16 promotes osteosarcoma progression and enhances cisplatin resistance by sponging miR-16 to upregulate ATG4B expression. Biochem. Biophys. Res. Commun. 2019, 518, 127–133. [Google Scholar] [CrossRef]
- Fennell, D. miR-16: Expanding the range of molecular targets in mesothelioma. Lancet Oncol. 2017, 18, 1296–1297. [Google Scholar] [CrossRef]
- Chen, Y.; Tsai, Y.H.; Tseng, B.J.; Pan, H.Y.; Tseng, S.H. Suppression of miR-19b enhanced the cytotoxic effects of mTOR inhibitors in human neuroblastoma cells. J. Pediatr. Surg. 2016, 51, 1818–1825. [Google Scholar] [CrossRef]
- Thorne, J.L.; Battaglia, S.; Baxter, D.E.; Hayes, J.L.; Hutchinson, S.A.; Jana, S.; Millican-Slater, R.A.; Smith, L.; Teske, M.C.; Wastall, L.M.; et al. MiR-19b non-canonical binding is directed by HuR and confers chemosensitivity through regulation of P-glycoprotein in breast cancer. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 996–1006. [Google Scholar] [CrossRef]
- Jiang, T.; Ye, L.; Han, Z.; Liu, Y.; Yang, Y.; Peng, Z.; Fan, J. miR-19b-3p promotes colon cancer proliferation and oxaliplatin-based chemoresistance by targeting SMAD4: Validation by bioinformatics and experimental analyses. J. Exp. Clin. Cancer Res. 2017, 36, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouvy, C.; Wannez, A.; Laloy, J.; Chatelain, C.; Dogné, J.M. Transfer of multidrug resistance among acute myeloid leukemia cells via extracellular vesicles and their microRNA cargo. Leuk. Res. 2017, 62, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Lu, F.Y.; Shi, R.H.; Feng, Y.D.; Zhao, X.D.; Lu, Z.P.; Xiao, L.; Zhou, G.Q.; Qiu, J.M.; Cheng, C.E. MiR-26b regulates 5-FU-resistance in human colorectal cancer via down-regulation of Pgp. Am. J. Cancer Res. 2018, 8, 2518–2527. [Google Scholar]
- Zhao, B.; Zhang, J.; Chen, X.; Xu, H.; Huang, B. Mir-26b inhibits growth and resistance to paclitaxel chemotherapy by silencing the CDC6 gene in gastric cancer. Arch. Med. Sci. 2019, 15, 498–503. [Google Scholar] [CrossRef] [Green Version]
- Tian, L.; Zhang, J.; Ren, X.; Liu, X.; Gao, W.; Zhang, C.; Sun, Y.; Liu, M. Overexpression of miR-26b decreases the cisplatin-esistance in laryngeal cancer by targeting ATF2. Oncotarget 2017, 8, 79023–79033. [Google Scholar] [CrossRef] [Green Version]
- Jin, F.; Wang, Y.; Li, M.; Zhu, Y.; Liang, H.; Wang, C.; Wang, F.; Zhang, C.Y.; Zen, K.; Li, L. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis. 2017, 8, e2540. [Google Scholar] [CrossRef]
- Zhao, N.; Wang, R.; Zhou, L.; Zhu, Y.; Gong, J.; Zhuang, S.M. MicroRNA-26b suppresses the NF-κB signaling and enhances the chemosensitivity of hepatocellular carcinoma cells by targeting TAK1 and TAB3. Mol. Cancer 2014, 13, 35. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Su, J.; Zhao, Z.; Hou, Y.; Yin, X.; Zheng, N.; Zhou, X.; Yan, J.; Xia, J.; Wang, Z. MiR-26b reverses temozolomide resistance via targeting Wee1 in glioma cells. Cell Cycle 2017, 16, 1954–1964. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.J.; Drakaki, A.; Iliopoulos, D.; Struhl, K. MiR-27b targets PPARγ to inhibit growth, tumor progression and the inflammatory response in neuroblastoma cells. Oncogene 2012, 31, 3818–3825. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Hua, X.; Qi, N.; Han, G.; Yu, J.; Yu, Y.; Wei, X.; Li, H.; Chen, X.; Leng, C.; et al. MiR-27b suppresses epithelial-mesenchymal transition and chemoresistance in lung cancer by targeting Snail1. Life Sci. 2020, 254, 117238. [Google Scholar] [CrossRef]
- Chen, D.; Si, W.; Shen, J.; Du, C.; Lou, W.; Bao, C.; Zheng, H.; Pan, J.; Zhong, G.; Xu, L.; et al. miR-27b-3p inhibits proliferation and potentially reverses multi-chemoresistance by targeting CBLB/GRB2 in breast cancer cells. Cell Death Dis. 2018, 9, 188. [Google Scholar] [CrossRef]
- Shang, Y.; Feng, B.; Zhou, L.; Ren, G.; Zhang, Z.; Fan, X.; Sun, Y.; Luo, G.; Liang, J.; Wu, K.; et al. The miR27b-CCNG1-P53-miR-508-5p axis regulates multidrug resistance of gastric cancer. Oncotarget 2016, 7, 538–549. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Cai, M.; Zhang, Y.; Tao, L.; Guo, R. miR-29c-3p inhibits autophagy and cisplatin resistance in ovarian cancer by regulating FOXP1/ATG14 pathway. Cell Cycle 2020, 19, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shou, H.; Wang, Q.; Liu, S. Investigation of the potential theranostic role of KDM5B/miR-29c signaling axis in paclitaxel resistant endometrial carcinoma. Gene 2019, 694, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, T.; Li, W.; Li, M.; Zuo, Q.; Zou, Q.; Xiao, B. The miR-29c-KIAA1199 axis regulates gastric cancer migration by binding with WBP11 and PTP4A3. Oncogene 2019, 38, 3134–3150. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.M.; Tang, B.F.; Li, Z.X.; Guo, H.B.; Cheng, J.L.; Song, P.P.; Zhao, X. MiR-29c reduces the cisplatin resistance of non-small cell lung cancer cells by negatively regulating the PI3K/Akt pathway. Sci. Rep. 2018, 8, 8007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, S.; Yang, Z.; Qiu, X.; Lv, R.; Liu, J.; Wu, M.; Liao, Y.; Liu, Q. miR-29c contribute to glioma cells temozolomide sensitivity by targeting O6-methylguanine-DNA methyltransferases indirectely. Oncotarget 2016, 7, 50229–50238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Li, Y.; Sun, J.; Wang, Q.; Sun, C.; Yan, Y.; Yu, L.; Cheng, D.; An, T.; Shi, C.; et al. Tumor-suppressive effects of miR-29c on gliomas. Neuroreport 2013, 24, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.J.; Sun, G.K.; Zhang, T.J.; Wu, D.H.; Zhou, J.D.; Ma, B.B.; Xu, Z.J.; Wen, X.M.; Chen, Q.; Yao, D.M.; et al. Down-regulation of miR-29c is a prognostic biomarker in acute myeloid leukemia and can reduce the sensitivity of leukemic cells to decitabine. Cancer Cell Int. 2019, 19, 177. [Google Scholar] [CrossRef]
- Mraz, M.; Malinova, K.; Kotaskova, J.; Pavlova, S.; Tichy, B.; Malcikova, J.; Stano Kozubik, K.; Smardova, J.; Brychtova, Y.; Doubek, M.; et al. miR-34a, miR-29c and miR-17-5p are downregulated in CLL patients with TP53 abnormalities. Leukemia 2009, 23, 1159–1163. [Google Scholar] [CrossRef] [Green Version]
- Ye, S.; Sun, B.; Wu, W.; Yu, C.; Tian, T.; Lian, Z.; Liang, Q.; Zhou, Y. LINC01123 facilitates proliferation, invasion and chemoresistance of colon cancer cells. Biosci. Rep. 2020, 40, BSR20194062. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Wang, J.J.; Yang, X.R.; Yu, Y.L. Upregulation of miR-34c after silencing E2F transcription factor 1 inhibits paclitaxel combined with cisplatin resistance in gastric cancer cells. World J. Gastroenterol. 2020, 26, 499–513. [Google Scholar] [CrossRef]
- Wu, H.; Huang, M.; Lu, M.; Zhu, W.; Shu, Y.; Cao, P.; Liu, P. Regulation of microtubule-associated protein tau (MAPT) by miR-34c-5p determines the chemosensitivity of gastric cancer to paclitaxel. Cancer Chemother. Pharmacol. 2013, 71, 1159–1171. [Google Scholar] [CrossRef]
- Xiao, S.; Li, Y.; Pan, Q.; Ye, M.; He, S.; Tian, Q.; Xue, M. MiR-34c/SOX9 axis regulates the chemoresistance of ovarian cancer cell to cisplatin-based chemotherapy. J. Cell Biochem. 2019, 120, 2940–2953. [Google Scholar] [CrossRef]
- Tung, S.L.; Huang, W.C.; Hsu, F.C.; Yang, Z.P.; Jang, T.H.; Chang, J.W.; Chuang, C.M.; Lai, C.R.; Wang, L.H. miRNA-34c-5p inhibits amphiregulin-induced ovarian cancer stemness and drug resistance via downregulation of the AREG-EGFR-ERK pathway. Oncogenesis 2017, 6, e326. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Jin, H.; Xu, C.X.; Bi, W.Z.; Wang, Y. MiR-34c inhibits osteosarcoma metastasis and chemoresistance. Med. Oncol. 2014, 31, 972. [Google Scholar] [CrossRef]
- Liu, C.; Hou, J.; Shan, F.; Wang, L.; Lu, H.; Ren, T. Long Non-Coding RNA CRNDE Promotes Colorectal Carcinoma Cell Progression and Paclitaxel Resistance by Regulating miR-126-5p/ATAD2 Axis. Onco Targets Ther. 2020, 13, 4931–4942. [Google Scholar] [CrossRef]
- Fu, R.; Tong, J.S. miR-126 reduces trastuzumab resistance by targeting PIK3R2 and regulating AKT/mTOR pathway in breast cancer cells. J. Cell. Mol. Med. 2020, 24, 7600–7608. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Chen, H.; Wong, N.; Haynes, W.; Baker, C.M.; Wang, X. Pseudohypoxia induced by miR-126 deactivation promotes migration and therapeutic resistance in renal cell carcinoma. Cancer Lett. 2017, 394, 65–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, M.J.; Raby, K.L.; Saini, H.K.; Bailey, S.; Wool, S.V.; Tunnacliffe, J.M.; Enright, A.J.; Nicholson, J.C.; Coleman, N. Solid tumors of childhood display specific serum microRNA profiles. Cancer Epidemiol. Biomark. Prev. 2015, 24, 350–360. [Google Scholar] [CrossRef] [Green Version]
- Wei, J.S.; Johansson, P.; Chen, Q.R.; Song, Y.K.; Durinck, S.; Wen, X.; Cheuk, A.T.; Smith, M.A.; Houghton, P.; Morton, C.; et al. microRNA profiling identifies cancer-specific and prognostic signatures in pediatric malignancies. Clin. Cancer Res. 2009, 15, 5560–5568. [Google Scholar] [CrossRef] [Green Version]
- Zeka, F.; Decock, A.; Van Goethem, A.; Vanderheyden, K.; Demuynck, F.; Lammens, T.; Helsmoortel, H.H.; Vermeulen, J.; Noguera, R.; Berbegall, A.P.; et al. Circulating microRNA biomarkers for metastatic disease in neuroblastoma patients. JCI Insight 2018, 3, e97021. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.K.; Lin, J.W.; Shih, J.W.; Chuang, H.Y.; Fong, I.H.; Yeh, C.T.; Lin, C.M. Targeting BC200/miR218-5p Signaling Axis for Overcoming Temozolomide Resistance and Suppressing Glioma Stemness. Cells 2020, 9, 1859. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, X.; Du, L.; Wang, Y.; Liu, X.; Tian, H.; Wang, L.; Li, P.; Zhao, Y.; Duan, W.; et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol. Cancer 2019, 18, 43. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zhan, M.; Xu, S.W.; Chen, W.; Long, M.M.; Shi, Y.H.; Liu, Q.; Mohan, M.; Wang, J. miR-218-5p restores sensitivity to gemcitabine through PRKCE/MDR1 axis in gallbladder cancer. Cell Death Dis. 2017, 8, e2770. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Yang, X.; Cheng, Y.; Zhang, X.; Yang, C.; Deng, X.; Li, P.; Tao, J.; Yang, H.; Wei, J.; et al. MicroRNA-218 Increases the Sensitivity of Bladder Cancer to Cisplatin by Targeting Glut1. Cell Physiol. Biochem. 2017, 41, 921–932. [Google Scholar] [CrossRef]
- Zarogoulidis, P.; Petanidis, S.; Kioseoglou, E.; Domvri, K.; Anestakis, D.; Zarogoulidis, K. MiR-205 and miR-218 expression is associated with carboplatin chemoresistance and regulation of apoptosis via Mcl-1 and Survivin in lung cancer cells. Cell. Signal. 2015, 27, 1576–1588. [Google Scholar] [CrossRef]
- Zeng, F.; Wang, Q.; Wang, S.; Liang, S.; Huang, W.; Guo, Y.; Peng, J.; Li, M.; Zhu, W.; Guo, L. Linc00173 promotes chemoresistance and progression of small cell lung cancer by sponging miR-218 to regulate Etk expression. Oncogene 2020, 39, 293–307. [Google Scholar] [CrossRef]
- Chen, X.; Pan, M.; Han, L.; Lu, H.; Hao, X.; Dong, Q. miR-338-3p suppresses neuroblastoma proliferation, invasion and migration through targeting PREX2a. FEBS Lett. 2013, 587, 3729–3737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Sun, Y.; Wang, D.; Sun, H.; Liu, X. SNHG16 promotes tumorigenesis and cisplatin resistance by regulating miR-338-3p/PLK4 pathway in neuroblastoma cells. Cancer Cell Int. 2020, 20, 236. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Cui, D.; Li, B.; Xu, W.W.; Lam, A.K.Y.; Chan, K.T.; Zhu, Y.; Lee, N.P.Y.; Law, S.Y.K.; Guan, X.Y.; et al. MicroRNA-338-5p reverses chemoresistance and inhibits invasion of esophageal squamous cell carcinoma cells by targeting Id-1. Cancer Sci. 2019, 110, 3677–3688. [Google Scholar] [CrossRef] [PubMed]
- Creevey, L.; Ryan, J.; Harvey, H.; Bray, I.M.; Meehan, M.; Khan, A.R.; Stallings, R.L. MicroRNA-497 increases apoptosis in MYCN amplified neuroblastoma cells by targeting the key cell cycle regulator WEE1. Mol. Cancer 2013, 12, 23. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Ji, X.B.; Wang, L.H.; Qiu, J.G.; Zhou, F.M.; Liu, W.J.; Wan, D.D.; Lin, M.C.; Liu, L.Z.; Zhang, J.Y.; et al. Regulation of MicroRNA-497-Targeting AKT2 Influences Tumor Growth and Chemoresistance to Cisplatin in Lung Cancer. Front. Cell Dev. Biol. 2020, 8, 840. [Google Scholar] [CrossRef]
- Wang, L.; Jiang, C.F.; Li, D.M.; Ge, X.; Shi, Z.M.; Li, C.Y.; Liu, X.; Yin, Y.; Zhen, L.; Liu, L.Z.; et al. MicroRNA-497 inhibits tumor growth and increases chemosensitivity to 5-fluorouracil treatment by targeting KSR1. Oncotarget 2016, 7, 2660–2671. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Fu, G.B.; Tao, Z.; Yang, J.O.; Kong, F.; Jiang, B.H.; Wan, X.; Chen, K. MiR-497 decreases cisplatin resistance in ovarian cancer cells by targeting mTOR/P70S6K1. Oncotarget 2015, 6, 26457–26471. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Wang, T.; Cao, Z.; Huang, H.; Li, J.; Liu, W.; Liu, S.; You, L.; Zhou, L.; Zhang, T.; et al. MiR-497 downregulation contributes to the malignancy of pancreatic cancer and associates with a poor prognosis. Oncotarget 2014, 5, 6983–6993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoareau-Aveilla, C.; Quelen, C.; Congras, A.; Caillet, N.; Labourdette, D.; Dozier, C.; Brousset, P.; Lamant, L.; Meggetto, F. miR-497 suppresses cycle progression through an axis involving CDK6 in ALK-positive cells. Haematologica 2019, 104, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Domingo-Fernandez, R.; Watters, K.; Piskareva, O.; Stallings, R.L.; Bray, I. The role of genetic and epigenetic alterations in neuroblastoma disease pathogenesis. Pediatr. Surg. Int. 2013, 29, 101–119. [Google Scholar] [CrossRef] [Green Version]
- Bedrnicek, J.; Vicha, A.; Jarosova, M.; Holzerova, M.; Cinatl, J., Jr.; Michaelis, M.; Cinatl, J.; Eckschlager, T. Characterization of drug-resistant neuroblastoma cell lines by comparative genomic hybridization. Neoplasma 2005, 52, 415–419. [Google Scholar] [PubMed]
- Wang, L.; Yang, G.; Zhao, D.; Wang, J.; Bai, Y.; Peng, Q.; Wang, H.; Fang, R.; Chen, G.; Wang, Z.; et al. CD103-positive CSC exosome promotes EMT of clear cell renal cell carcinoma: Role of remote MiR-19b-3p. Mol. Cancer 2019, 18, 86. [Google Scholar] [CrossRef] [PubMed]
- Peng, P.; Chen, T.; Wang, Q.; Zhang, Y.; Zheng, F.; Huang, S.; Tang, Y.; Yang, C.; Ding, W.; Ren, D.; et al. Decreased miR-218-5p Levels as a Serum Biomarker in Bone Metastasis of Prostate Cancer. Oncol. Res. Treat. 2019, 42, 165–185. [Google Scholar] [CrossRef]
- Ahmadinejad, F.; Mowla, S.J.; Honardoost, M.A.; Arjenaki, M.G.; Moazeni-Bistgani, M.; Kheiri, S.; Teimori, H. Lower expression of miR-218 in human breast cancer is associated with lymph node metastases, higher grades, and poorer prognosis. Tumour Biol. 2017, 39. [Google Scholar] [CrossRef] [Green Version]
- Deng, M.; Zeng, C.; Lu, X.; He, X.; Zhang, R.; Qiu, Q.; Zheng, G.; Jia, X.; Liu, H.; He, Z. miR-218 suppresses gastric cancer cell cycle progression through the CDK6/Cyclin D1/E2F1 axis in a feedback loop. Cancer Lett. 2017, 403, 175–185. [Google Scholar] [CrossRef]
- Jiang, Z.; Song, Q.; Zeng, R.; Li, J.; Li, J.; Lin, X.; Chen, X.; Zhang, J.; Zheng, Y. MicroRNA-218 inhibits EMT, migration and invasion by targeting SFMBT1 and DCUN1D1 in cervical cancer. Oncotarget 2016, 7, 45622–45636. [Google Scholar] [CrossRef] [Green Version]
- Chiu, K.L.; Kuo, T.T.; Kuok, Q.Y.; Lin, Y.S.; Hua, C.H.; Lin, C.Y.; Su, P.Y.; Lai, L.C.; Sher, Y.P. ADAM9 enhances CDCP1 protein expression by suppressing miR-218 for lung tumor metastasis. Sci. Rep. 2015, 5, 16426. [Google Scholar] [CrossRef] [Green Version]
- Marengo, B.; Pulliero, A.; Izzotti, A.; Domenicotti, C. miRNA Regulation of Glutathione Homeostasis in Cancer Initiation, Progression and Therapy Resistance. Microrna 2020, 9, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Stigliani, S.; Scaruffi, P.; Lagazio, C.; Persico, L.; Carlini, B.; Varesio, L.; Morandi, F.; Morini, M.; Gigliotti, A.R.; Esposito, M.R.; et al. Deregulation of focal adhesion pathway mediated by miR-659-3p is implicated in bone marrow infiltration of stage M neuroblastoma patients. Oncotarget 2015, 6, 13295–13308. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| miRNA | HTLA-230/ER-HTLA Ratio | Expression in Neuroblastoma | Expression in Other Chemoresistant Cancers |
|---|---|---|---|
| miR-15a | 11.38 | Down-regulated in MYCN-amplified chemoresistant NB [13,22,23,24] | Down-regulated in Burkitt Lymphoma [25], pancreatic ductal adenocarcinoma [26], colorectal [27], and ovarian cancer [28] |
| miR-16 | 7.89 | Down-regulated in MYCN-amplified NB [22,29] and in chemoresistant NB [13] | Down-regulated in cervical [30], breast [31,32] gastric [32,33], and lung [34] cancer, osteosarcoma [35], and mesothelioma [36] |
| miR-19b | 7.75 | Up-regulated in chemoresistant NB [37] | Down-regulated in breast [38] and colon [39] cancer and leukemia [40] |
| miR-26b | 47.87 | Not evaluated | Down-regulated in chemoresistant colorectal [41], gastric [42], laryngeal [43], and hepatocellular carcinoma [44,45] cancer and in glioma [46] |
| miR-27b | 8.29 | Down-regulated in NB [47] | Down-regulated in lung [48], breast [49], and gastric cancer [50] |
| miR-29c | 9.27 | Not evaluated | Down-regulated in ovarian [51], endometrial [52], gastric [53], and small cell lung [54] cancer, glioma [55,56], and leukemia [57,58] |
| miR-34c | 7.49 | Not evaluated | Down-regulated in colon [59], gastric [60,61], and ovarian [62,63] cancer, and osteosarcoma [64] |
| miR-126a | 9.66 | Not evaluated | Down-regulated in colorectal [65] and breast cancer [66] and in renal cell carcinoma [67] |
| miR-218 | 12.30 | Up-regulated in MYCN-amplified and in metastatic NB [68,69,70] | Down-regulated in glioma cells [71], colorectal [72], gallbladder [73], bladder [74], and lung cancer [75,76] |
| miR-338 | 15.99 | Down-regulated in resistant NB [77,78] | Down-regulated in esophageal squamous carcinoma cells [79] |
| miR-497 | 6.92 | Down-regulated in chemoresistant NB [24], in MYCN-amplified NB [80] | Down-regulated in lung [81], colorectal [82], ovarian [83], and pancreatic [84] cancer, and lymphoma [85] |
| CHR | START | STOP | CYTO | SIZE KB | VALUE | Control (HTLA-230) |
|---|---|---|---|---|---|---|
| 1 | 152,079,488 | 155,154,990 | q21.3–q22 | 3.075 | 1.5 | Absent |
| 3 | 73,792,065 | 75,028,724 | p13–p12.3 | 1.236 | −0.7 | Absent |
| 5 | 20,160,410 | 44,924,503 | p14.3–p12 | 24.764 | −0.7 | Absent |
| 8 | 112,697,432 | 146,280,020 | q23.3–q24.3 | 33.582 | −1/−0.4 | Duplicated |
| 9 | 204,193 | 38,815,475 | p24.3–p13.1 | 38.611 | −0.7/−3 | Absent |
| 10 | 43,020,732 | 60,914,512 | q11.21–q21.1 | 17.893 | −0.7/−1.2 | Duplicated |
| 12 | 38,805,636 | 48,103,580 | q12–q13.11 | 9.297 | 0.7/0.4/1.4 | Absent |
| 13 | 20,412,619 | 39,841,779 | q12.11–q13.3 | 19.429 | 0.3/0.6 | Absent |
| 13 | 39,900,189 | 86,110,407 | q13.3–q31.1 | 46.210 | −0.5 | Absent |
| 13 | 86,151,801 | 111,106,213 | q31.1–q34 | 24.954 | 0.5 | Absent |
| 13 | 111,181,035 | 113,538,619 | q34 | 2.357 | −0.8 | Absent |
| 13 | 113,610,612 | 115,092,648 | q34 | 1.482 | 0.4 | Absent |
| 17 | 44,684 | 625,475 | p13.3 | 580 | −0.7 | Absent |
| 17 | 25,654,874 | 40,109,636 | q11.1–q21.2 | 14.454 | −0.7/−1.2 | Duplicated |
| 19 | 32,783,771 | 36,293,337 | q13.11–q13.12 | 3.509 | −0.6 | Duplicated |
| 20 | 60,747 | 19,483,849 | p13–p11.23 | 19.423 | 0.5 | Deleted or mosaic |
| 21 | 15,538,980 | 32,776,404 | q11.2–q22.11 | 17.237 | −0.6 | Absent |
| N | MYCN Status | Age (Months) | EFS (Months) | OS (Months) | INRG Stage | Induction Response | Relapse | Follow-Up |
|---|---|---|---|---|---|---|---|---|
| 2 | Amplified | 55 | 71.25 | 71.25 | M | Yes | No | Alive |
| 3 | Not evaluated | 41 | 81.06 | 81.06 | M | Yes | No | Alive |
| 4 | Single copy | 12 | 60.53 | 60.53 | M | Yes | No | Alive |
| 6 | Amplified | 62 | 50.83 | 50.83 | M | Yes | No | Alive |
| 9 | Amplified | 21 | 55.38 | 55.38 | M | Yes | No | Alive |
| 1 | Amplified | 17 | 6.86 | 7.10 | M | No | Yes | Dead |
| 5 | Single copy | 47 | 22.94 | 26.17 | M | No | Yes | Dead |
| 7 | Amplified | 20 | 9.54 | 19.27 | M | No | Yes | Dead |
| 8 | Amplified | 21 | 5.54 | 7.00 | M | No | Yes | Dead |
| 10 | Amplified | 16 | 4.69 | 8.98 | M | No | Yes | Dead |
| N | MYCN Status | Age (Years) | EFS (Months) | OS (Months) | |
|---|---|---|---|---|---|
| Tumors | 1 | Amplified | 1.99 | 46.2 | 84.8 |
| 2 | Not amplified | 3.18 | 5.3 | 9.3 | |
| 3 | Not amplified | 1.27 | 187.2 | 187.2 | |
| 4 | Amplified | 1.13 | 4.3 | 7.4 | |
| 5 | Amplified | 3.88 | 70.0 | 114.5 | |
| 6 | Amplified | 6.30 | 36.2 | 47.7 | |
| 7 | Amplified | 2.07 | 3.2 | 10.3 | |
| 8 | Amplified | 4.76 | 114,9 | 114,9 | |
| 9 | Amplified | 4.57 | 21.7 | 22.4 | |
| 10 | Amplified | 1.44 | 26.8 | 33.0 | |
| Metastases | 11 | Not amplified | 1.68 | 58.88 | 58.88 |
| 12 | Not amplified | 3 | 24.52 | 35.98 | |
| 13 | Amplified | 6.8 | 8.68 | 42.41 | |
| 14 | Amplified | 0.9 | 11.06 | 11.06 | |
| 15 | Not amplified | 2.55 | 70.73 | 70.73 | |
| 16 | Not amplified | 3.34 | 13,3 | 21.22 | |
| 17 | Amplified | 8.2 | 8.61 | 8.61 | |
| 18 | Amplified | 1.67 | 9.6 | 14.98 | |
| 19 | Amplified | 6.89 | 29.04 | 29.04 | |
| 20 | Amplified | 1.7 | 13.3 | 23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marengo, B.; Pulliero, A.; Corrias, M.V.; Leardi, R.; Farinini, E.; Fronza, G.; Menichini, P.; Monti, P.; Monteleone, L.; Valenti, G.E.; et al. Potential Role of miRNAs in the Acquisition of Chemoresistance in Neuroblastoma. J. Pers. Med. 2021, 11, 107. https://doi.org/10.3390/jpm11020107
Marengo B, Pulliero A, Corrias MV, Leardi R, Farinini E, Fronza G, Menichini P, Monti P, Monteleone L, Valenti GE, et al. Potential Role of miRNAs in the Acquisition of Chemoresistance in Neuroblastoma. Journal of Personalized Medicine. 2021; 11(2):107. https://doi.org/10.3390/jpm11020107
Chicago/Turabian StyleMarengo, Barbara, Alessandra Pulliero, Maria Valeria Corrias, Riccardo Leardi, Emanuele Farinini, Gilberto Fronza, Paola Menichini, Paola Monti, Lorenzo Monteleone, Giulia Elda Valenti, and et al. 2021. "Potential Role of miRNAs in the Acquisition of Chemoresistance in Neuroblastoma" Journal of Personalized Medicine 11, no. 2: 107. https://doi.org/10.3390/jpm11020107