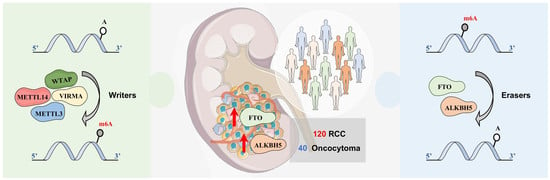
Deregulation of N6-Methyladenosine RNA Modification and Its Erasers FTO/ALKBH5 among the Main Renal Cell Tumor Subtypes
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Silico Analysis
2.2. Patients and Samples
2.3. RNA Extraction, cDNA Synthesis and RT-qPCR
2.4. Immunohistochemical Analysis
2.5. Statistical Analysis
3. Results
3.1. In Silico Analysis of m6A-Related Proteins in TCGA’s RCC Patients
3.2. Differential FTO and ALKBH5 Expression among RCC Subtypes and Oncocytoma in IPOPorto’s Cohort
3.3. Evaluation of FTO, ALKBH5 and m6A Immunoexpression in Primary Tumors
3.4. Association with Clinicopathological Parameters and Chromosomal Aberrations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AJCC | American Joint Committee on Cancer |
| AUC | area under the curve |
| ccRCC | clear cell renal cell carcinoma |
| cDNA | complementary DNA |
| chRCC | chromophobe renal cell carcinoma |
| FFPE | formalin-fixed paraffin-embedded |
| GUSB | beta-glucoronidase |
| IHC | immunohistochemistry |
| m6A | N-6-methyladenosine |
| mRNA | messenger RNA |
| OR | odds ratio |
| pRCC | papillary renal cell carcinoma |
| PPV | positive predictive value |
| RNA | ribonucleic acid |
| ROC | receiver operating characteristics |
| rs | Spearman’s correlation coefficient |
| RT-qPCR | real-time quantitative polymerase chain reaction |
| TCGA | The Cancer Genome Atlas |
| WHO | World Health Organization |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clinicians 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Haake, S.M.; Rathmell, W.K. Renal cancer subtypes: Should we be lumping or splitting for therapeutic decision making? Cancer 2017, 123, 200–209. [Google Scholar] [CrossRef]
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Primers 2017, 3, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.M.; Lynch, D.T. Renal Oncocytoma; StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Linehan, W.M.; Ricketts, C.J. The Cancer Genome Atlas of renal cell carcinoma: Findings and clinical implications. Nat. Rev. Urology 2019, 16, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Padala, S.A.; Barsouk, A.; Thandra, K.C.; Saginala, K.; Mohammed, A.; Vakiti, A.; Rawla, P.; Barsouk, A. Epidemiology of Renal Cell Carcinoma. World J. Oncol. 2020, 11, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Keizman, D.; Gottfriedm, M.; Ish-Shalom, M.; Maimon, N.; Peer, A.; Neumann, A.; Hammers, H.; Eisenberger, M.A.; Sinibaldi, V.; Pili, R.; et al. Active smoking may negatively affect response rate, progression-free survival, and overall survival of patients with metastatic renal cell carcinoma treated with sunitinib. Oncologist 2014, 19, 51–60. [Google Scholar] [CrossRef] [Green Version]
- Lobo, J.; Barros-Silva, D.; Henrique, R.; Jeronimo, C. The Emerging Role of Epitranscriptomics in Cancer: Focus on Urological Tumors. Genes 2018, 9, 552. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Qi, X.; Liu, L.; Ma, S.; Liu, J.; Wu, J. Epigenetic Regulation of m(6)A Modifications in Human Cancer. Mol. Ther. Nucleic Acids 2020, 19, 405–412. [Google Scholar] [CrossRef]
- Lan, Q.; Liu, P.Y.; Haase, J.; Bell, J.L.; Huttelmaier, S.; Liu, T. The Critical Role of RNA m(6)A Methylation in Cancer. Cancer Res. 2019, 79, 1285–1292. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Wei, J.; Cui, Y.H.; Park, G.; Shah, P.; Deng, Y.; Aplin, A.E.; Lu, Z.; Hwang, S.; He, C.; et al. M(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 2019, 10, 2782. [Google Scholar] [CrossRef]
- Zhu, W.; Wang, J.Z.; Xu, Z.; Cao, M.; Hu, Q.; Pan, C.; Guo, M.; Wei, J.F.; Yang, H. Detection of N6methyladenosine modification residues (Review). Int. J. Mol. Med. 2019, 43, 2267–2278. [Google Scholar]
- Wang, T.; Kong, S.; Tao, M.; Ju, S. The potential role of RNA N6-methyladenosine in Cancer progression. Mol. Cancer 2020, 19, 88. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, B.; Nie, Z.; Duan, L.; Xiong, Q.; Jin, Z.; Yang, C.; Chen, Y. The role of m6A modification in the biological functions and diseases. Signal. Transduct. Target. Ther. 2021, 6, 74. [Google Scholar] [CrossRef]
- Ma, J.Z.; Yang, F.; Zhou, C.C.; Liu, F.; Yuan, J.H.; Wang, F.; Wang, T.T.; Xu, Q.G.; Zhou, W.P.; Sun, S.H. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology 2017, 65, 529–543. [Google Scholar] [CrossRef]
- Cui, Q.; Shi, H.; Ye, P.; Li, L.; Qu, Q.; Sun, G.; Sun, G.; Lu, Z.; Huang, Y.; Yang, C.G.; et al. M(6)A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017, 18, 2622–2634. [Google Scholar] [CrossRef]
- Weng, H.; Huang, H.; Wu, H.; Qin, X.; Zhao, B.S.; Dong, L.; Shi, H.; Skibbe, J.; Shen, C.; Hu, C.; et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m(6)A Modification. Cell Stem cell 2018, 22, 191–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barros-Silva, D.; Lobo, J.; Guimarães-Teixeira, C.; Carneiro, I.; Oliveira, J.; Martens-Uzunova, E.S.; Henrique, R.; Jerónimo, C. VIRMA-Dependent N6-Methyladenosine Modifications Regulate the Expression of Long Non-Coding RNAs CCAT1 and CCAT2 in Prostate Cancer. Cancers 2020, 12, 771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lobo, J.; Costa, A.L.; Cantante, M.; Guimaraes, R.; Lopes, P.; Antunes, L.; Braga, I.; Oliveira, J.; Pelizzola, M.; Henrique, R.; et al. M(6)A RNA modification and its writer/reader VIRMA/YTHDF3 in testicular germ cell tumors: A role in seminoma phenotype maintenance. J. Transl. Med. 2019, 17, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; Wu, D.; Ning, J.; Liu, W.; Zhang, D. Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer 2019, 19, 326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ianniello, Z.; Fatica, A. N6-Methyladenosine Role in Acute Myeloid Leukaemia. Int. J. Mol. Sci. 2018, 19, 2345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Cong, R.; Liu, S.; Zhu, B.; Wang, X.; Xing, Q. Decreased expression of METTL14 predicts poor prognosis and construction of a prognostic signature for clear cell renal cell carcinoma. Cancer Cell Int. 2021, 21, 46. [Google Scholar] [CrossRef]
- Li, X.; Tang, J.; Huang, W.; Wang, F.; Li, P.; Qin, C.; Qin, Z.; Zou, Q.; Wei, J.; Hua, L.; et al. The M6A methyltransferase METTL3: Acting as a tumor suppressor in renal cell carcinoma. Oncotarget 2017, 8, 96103–96116. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Yu, K.; Zhong, G.; Shen, W. Identification of a m(6)A RNA methylation regulators-based signature for predicting the prognosis of clear cell renal carcinoma. Cancer Cell Int. 2020, 20, 157. [Google Scholar] [CrossRef]
- Strick, A.; Von Hagen, F.; Gundert, L.; Klümper, N.; Tolkach, Y.; Schmidt, D.; Kristiansen, G.; Toma, M.; Ritter, M.; Ellinger, J. The N(6) -methyladenosine (m(6) A) erasers alkylation repair homologue 5 (ALKBH5) and fat mass and obesity-associated protein (FTO) are prognostic biomarkers in patients with clear cell renal carcinoma. BJU Int. 2020, 125, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.J.; Luan, J.C.; Song, L.B.; Cong, R.; Ji, C.J.; Zhou, X.; Xia, J.D.; Song, N.H. m6A RNA methylation regulators correlate with malignant progression and have potential predictive values in clear cell renal cell carcinoma. Exp. Cell Res. 2020, 392, 112015. [Google Scholar] [CrossRef]
- Zhang, Y.; Yao, Y.; Qi, X.; Li, J.; Liu, M.; Che, X.; Xu, Y.; Wu, G. Identification of a New Prognostic Risk Signature of Clear Cell Renal Cell Carcinoma Based on N(6)-Methyladenosine RNA Methylation Regulators. J. Immunol. Res. 2021, 2021, 6617841. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Mao, S.; Guo, Y.; Zhang, W.; Liu, J.; Li, C.; Yao, X. N6-methyladenosine RNA methylation regulators participate in malignant progression and have prognostic value in clear cell renal cell carcinoma. Oncol. Rep. 2020, 43, 1591–1605. [Google Scholar] [CrossRef] [Green Version]
- Zhong, J.; Liu, Z.; Cai, C.; Duan, X.; Deng, T.; Zeng, G. M(6)A modification patterns and tumor immune landscape in clear cell renal carcinoma. J. Immunother. Cancer 2021, 9, e001646. [Google Scholar] [CrossRef]
- Green, N.H.; Galvan, D.L.; Badal, S.S.; Chang, B.H.; LeBleu, V.S.; Long, J.; Jonasch, E.; Danesh, F.R. MTHFD2 links RNA methylation to metabolic reprogramming in renal cell carcinoma. Oncogene 2019, 38, 6211–6225. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, F.; Wang, Z.; Yang, X.; Yu, H.; Si, S.; Lu, J.; Zhou, Z.; Lu, Q.; Wang, Z.; et al. ALKBH5 promotes the proliferation of renal cell carcinoma by regulating AURKB expression in an m(6)A-dependent manner. Ann. Transl. Med. 2020, 8, 646. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, C.; Zhuang, C.; Luo, X.; Huang, X.; Yao, L.; Li, J.; Li, Y.; Xiong, T.; Ye, J.; Zhang, F.; et al. N6-methyladenosine demethylase FTO suppresses clear cell renal cell carcinoma through a novel FTO-PGC-1α signalling axis. J. Cell Mol. Med. 2019, 23, 2163–2173. [Google Scholar] [CrossRef]
- Kang, H.; Zhang, Z.; Yu, L.; Li, Y.; Liang, M.; Zhou, L. FTO reduces mitochondria and promotes hepatic fat accumulation through RNA demethylation. J. Cell Biochem. 2018, 119, 5676–5685. [Google Scholar] [CrossRef]
- Marquardt, A.; Solimando, A.G.; Kerscher, A.; Bittrich, M.; Kalogirou, C.; Kübler, H.; Rosenwald, A.; Bargou, R.; Kollmannsberger, P.; Schilling, B.; et al. Subgroup-Independent Mapping of Renal Cell Carcinoma-Machine Learning Reveals Prognostic Mitochondrial Gene Signature Beyond Histopathologic Boundaries. Front. Oncol. 2021, 11, 621278. [Google Scholar] [CrossRef]
- Moch, H.; Ulbright, T.; Humphrey, P.; Reuter, V. WHO Classification of Tumours of the Urinary System and Male Genital Organs, 4th ed.; IARC: Lyon, France, 2016. [Google Scholar]
- The American Joint Commission on Cancer. AJCC Cancer Staging Manual; Springer International Publishing: Basel, Switzerland, 2017. [Google Scholar]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieira, J.; Henrique, R.; Ribeiro, F.R.; Barros-Silva, J.D.; Peixoto, A.; Santos, C.; Pinheiro, M.; Costa, V.L.; Soares, M.J.; Oliveira, J.; et al. Feasibility of differential diagnosis of kidney tumors by comparative genomic hybridization of fine needle aspiration biopsies. Genes Chromosomes Cancer 2010, 49, 935–947. [Google Scholar] [CrossRef] [PubMed]
- Jonasch, E.; Gao, J.; Rathmell, W.K. Renal cell carcinoma. BMJ 2014, 349, g4797. [Google Scholar] [CrossRef] [PubMed]
- Mitomi, T.; Kawahara, T.; Nomura, S.; Kuroda, S.; Takeshima, T.; Takamoto, D.; Otani, M.; Uemura, H. Skin Metastasis of Renal Cell Carcinoma. Case Rep. Oncol. 2020, 13, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Yang, J.; Huang, Q.; Jiang, M.J.; Tan, Y.N.; Fu, J.F.; Zhu, L.Z.; Fang, X.F.; Yuan, Y. Different treatment strategies and molecular features between right-sided and left-sided colon cancers. World J. Gastroenterol. WJG 2015, 21, 6470–6478. [Google Scholar] [CrossRef]
- Wiener, D.; Schwartz, S. The epitranscriptome beyond m(6)A. Nat. Rev. Genet. 2021, 22, 119–131. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef] [Green Version]
- Peer, E.; Moshitch-Moshkovitz, S.; Rechavi, G.; Dominissini, D. The Epitranscriptome in Translation Regulation. Cold Spring Harb Perspect. Biol. 2019, 11, a032623. [Google Scholar] [CrossRef]
- Sun, T.; Wu, R.; Ming, L. The role of m6A RNA methylation in cancer. Biomed. Pharmacother. 2019, 112, 108613. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Wei, J.; He, C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef]
- Dango, S.; Mosammaparast, N.; Sowa, M.E.; Xiong, L.J.; Wu, F.; Park, K.; Rubin, M.; Gygi, S.; Harper, J.W.; Shi, Y. DNA unwinding by ASCC3 helicase is coupled to ALKBH3-dependent DNA alkylation repair and cancer cell proliferation. Mol. Cell 2011, 44, 373–384. [Google Scholar] [CrossRef] [Green Version]
- Fedeles, B.I.; Singh, V.; Delaney, J.C.; Li, D.; Essigmann, J.M. The AlkB Family of Fe(II)/α-Ketoglutarate-dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond. J. Biol. Chem. 2015, 290, 20734–20742. [Google Scholar] [CrossRef] [Green Version]
- Tao, L.; Mu, X.; Chen, H.; Jin, D.; Zhang, R.; Zhao, Y.; Fan, J.; Cao, M.; Zhou, Z. FTO modifies the m6A level of MALAT and promotes bladder cancer progression. Clin. Transl. Med. 2021, 11, e310. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.M.; Batista, P.J.; Meier, J.L. Metabolic Regulation of the Epitranscriptome. ACS Chem. Biol. 2019, 14, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Su, R.; Stanford, S.; Chen, J. Critical Enzymatic Functions of FTO in Obesity and Cancer. Front. Endocrinol. 2018, 9, 396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Zhang, Z. ALKBH5-mediated m6A demethylation of lncRNA RMRP plays an oncogenic role in lung adenocarcinoma. Mamm. Genome 2021, 32, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Guo, S.; Piao, H.Y.; Wang, Y.; Wu, Y.; Meng, X.Y.; Yang, D.; Zheng, Z.C.; Zhao, Y. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J. Physiol. Biochem. 2019, 75, 379–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Z.; Wang, X.; Xu, Z.; Cao, Y.; Gong, R.; Yu, Y.; Yu, Y.; Guo, X.; Liu, S.; Yu, M.; et al. ALKBH5 regulates cardiomyocyte proliferation and heart regeneration by demethylating the mRNA of YTHDF1. Theranostics 2021, 11, 3000–3016. [Google Scholar] [CrossRef]
- Huff, S.; Tiwari, S.K.; Gonzalez, G.M.; Wang, Y.; Rana, T.M. M(6)A-RNA Demethylase FTO Inhibitors Impair Self-Renewal in Glioblastoma Stem Cells. ACS Chem. Biol. 2021, 16, 324–333. [Google Scholar] [CrossRef]
- Malacrida, A.; Rivara, M.; Di Domizio, A.; Cislaghi, G.; Miloso, M.; Zuliani, V.; Nicolini, G. 3D proteome-wide scale screening and activity evaluation of a new ALKBH5 inhibitor in U87 glioblastoma cell line. Bioorg. Med. Chem. 2020, 28, 115300. [Google Scholar] [CrossRef]
- Li, J.; Han, Y.; Zhang, H.; Qian, Z.; Jia, W.; Gao, Y.; Zheng, H.; Li, B. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem. Biophys. Res. Commun. 2019, 512, 479–485. [Google Scholar] [CrossRef]
- Niu, Y.; Lin, Z.; Wan, A.; Chen, H.; Liang, H.; Sun, L.; Wang, Y.; Li, X.; Xiong, X.F.; Wei, B.; et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol. Cancer 2019, 18, 46. [Google Scholar] [CrossRef] [Green Version]
- Zou, D.; Dong, L.; Li, C.; Yin, Z.; Rao, S.; Zhou, Q. The m(6)A eraser FTO facilitates proliferation and migration of human cervical cancer cells. Cancer Cell Int. 2019, 19, 321. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.; Sheng, Y.; Zhu, A.C.; Robinson, S.; Jiang, X.; Dong, L.; Chen, H.; Su, R.; Yin, Z.; Li, W.; et al. RNA Demethylase ALKBH5 Selectively Promotes Tumorigenesis and Cancer Stem Cell Self-Renewal in Acute Myeloid Leukemia. Cell Stem Cell 2020, 27, 64–80. [Google Scholar] [CrossRef]
- Thomsen, M.B.H.; Nordentoft, I.; Lamy, P.; Vang, S.; Reinert, L.; Mapendano, C.K.; Høyer, S.; Ørntoft, T.F.; Jensen, J.B.; Dyrskjøt, L. Comprehensive multiregional analysis of molecular heterogeneity in bladder cancer. Sci. Rep. 2017, 7, 11702. [Google Scholar] [CrossRef] [Green Version]
- Aran, D.; Camarda, R.; Odegaard, J.; Paik, H.; Oskotsky, B.; Krings, G.; Goga, A.; Sirota, M.; Butte, A.J. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat. Commun. 2017, 8, 1077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Y.; Thakkar, K.N.; Zhao, H.; Broughton, J.; Li, Y.; Seoane, J.A.; Diep, A.N.; Metzner, T.J.; Von Eyben, R.; Dill, D.L.; et al. The m(6)A RNA demethylase FTO is a HIF-independent synthetic lethal partner with the VHL tumor suppressor. Proc. Natl. Acad. Sci. USA 2020, 117, 21441–21449. [Google Scholar] [CrossRef] [PubMed]



| Progression-Free Survival (PFS) | HR | 95% CI for HR | p-Value | HR | 95% CI for HR | p-Value | |
|---|---|---|---|---|---|---|---|
| Variable | ccRCC | pRCC | |||||
| FTO expression | ≤P25 | 1.00 | - | - | 1.00 | - | - |
| >P25 | 0.67 | 0.43–1.05 | 0.081 | 0.42 | 0.24–0.75 | 0.004 | |
| Pathological stage | pT1 | 1.00 | - | - | 1.00 | - | - |
| pT2 | 3.08 | 1.59–5.95 | <0.001 | 3.58 | 1.59–8.04 | 0.020 | |
| pT3/4 | 5.96 | 3.54–10.05 | <0.001 | 7.18 | 3.82–13.50 | <0.001 | |
| Overall Survival (OS) | HR | 95% CI for HR | p-Value | HR | 95% CI for HR | p-Value | |
| Variable | ccRCC | pRCC | |||||
| FTO expression | ≤P25 | 1.00 | - | 0.035 | 1.00 | - | - |
| >P25 | 0.62 | 0.40–0.97 | <0.001 | 0.48 | 0.24–0.97 | 0.040 | |
| Age at diagnosis | - | 1.05 | 1.03–1.07 | <0.001 | - | - | - |
| Pathological stage | pT1 | 1.00 | - | - | 1.00 | - | - |
| pT2 | 1.25 | 0.62–2.53 | 0.5298 | 3.34 | 1.21–9.21 | 0.020 | |
| pT3/4 | 3.24 | 2.04–5.13 | <0.001 | 8.25 | 3.82–17.79 | <0.001 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guimarães-Teixeira, C.; Barros-Silva, D.; Lobo, J.; Soares-Fernandes, D.; Constâncio, V.; Leite-Silva, P.; Silva-Santos, R.; Braga, I.; Henrique, R.; Miranda-Gonçalves, V.; et al. Deregulation of N6-Methyladenosine RNA Modification and Its Erasers FTO/ALKBH5 among the Main Renal Cell Tumor Subtypes. J. Pers. Med. 2021, 11, 996. https://doi.org/10.3390/jpm11100996
Guimarães-Teixeira C, Barros-Silva D, Lobo J, Soares-Fernandes D, Constâncio V, Leite-Silva P, Silva-Santos R, Braga I, Henrique R, Miranda-Gonçalves V, et al. Deregulation of N6-Methyladenosine RNA Modification and Its Erasers FTO/ALKBH5 among the Main Renal Cell Tumor Subtypes. Journal of Personalized Medicine. 2021; 11(10):996. https://doi.org/10.3390/jpm11100996
Chicago/Turabian StyleGuimarães-Teixeira, Catarina, Daniela Barros-Silva, João Lobo, Diana Soares-Fernandes, Vera Constâncio, Pedro Leite-Silva, Rui Silva-Santos, Isaac Braga, Rui Henrique, Vera Miranda-Gonçalves, and et al. 2021. "Deregulation of N6-Methyladenosine RNA Modification and Its Erasers FTO/ALKBH5 among the Main Renal Cell Tumor Subtypes" Journal of Personalized Medicine 11, no. 10: 996. https://doi.org/10.3390/jpm11100996