Noncoding RNAs and Liquid Biopsy in Lung Cancer: A Literature Review
Abstract
:1. Introduction
2. Molecular Genetics of Lung Tumors
3. Noncoding RNAs in Lung Cancer
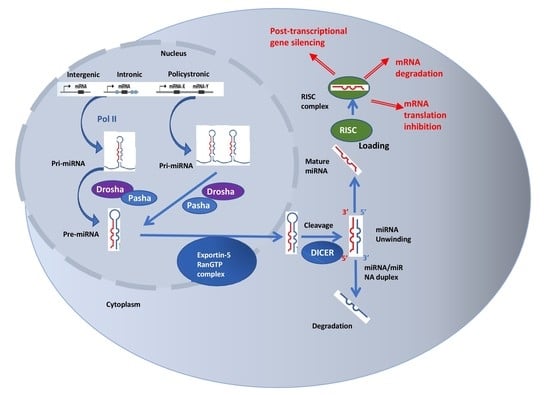
4. MicroRNAs
5. Long Noncoding RNAs
6. Liquid Biopsy in Lung Cancer
7. Circulating Noncoding RNAs
8. Detecting EGFR, ALK, and ROS1 in Plasma
9. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Sanger, F.; Coulson, A.R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 1975, 94, 441–448. [Google Scholar] [CrossRef]
- Stratton, M.R.; Campbell, P.J.; Futreal, P.A. The cancer genome. Nature 2009, 458, 719–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowell, P.C. The clonal evolution of tumor cell populations. Science 1976, 194, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Getz, G.; Wheeler, D.A.; Mardis, E.R.; McLellan, M.D.; Cibulskis, K.; Sougnez, C.; Greulich, H.; Muzny, D.M.; Morgan, M.B.; et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008, 455, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Behjati, S.; Tarpey, P.S. What is next generation sequencing? Arch. Dis. Child. Educ. Pract. Ed. 2013, 98, 236–238. [Google Scholar] [CrossRef]
- Campbell, P.J.; Stephens, P.J.; Pleasance, E.D.; O’Meara, S.; Li, H.; Santarius, T.; Stebbings, L.A.; Leroy, C.; Edkins, S.; Hardy, C.; et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 2008, 40, 722–729. [Google Scholar] [CrossRef]
- Berindan-Neagoe, I.; Chiorean, R.; Braicu, C.; Florian, I.S.; Leucuta, D.; Crisan, D.; Cocis, A.; Balacescu, O.; Irimie, A. Quantitative mRNA expression of genes involved in angiogenesis, coagulation and inflammation in multiforme glioblastoma tumoral tissue versus peritumoral brain tissue: Lack of correlation with clinical data. Eur. Cytokine Netw. 2012, 23, 45–55. [Google Scholar] [CrossRef] [Green Version]
- Ordway, J.M.; Budiman, M.A.; Korshunova, Y.; Maloney, R.K.; Bedell, J.A.; Citek, R.W.; Bacher, B.; Peterson, S.; Rohlfing, T.; Hall, J.; et al. Identification of novel high-frequency DNA methylation changes in breast cancer. PLoS ONE 2007, 2, e1314. [Google Scholar] [CrossRef] [Green Version]
- Sugarbaker, D.J.; Richards, W.G.; Gordon, G.J.; Dong, L.; Rienzo, A.; Maulik, G.; Glickman, J.N.; Chirieac, L.R.; Hartman, M.L.; Taillon, B.E.; et al. Transcriptome sequencing of malignant pleural mesothelioma tumors. Proc. Natl. Acad. Sci. USA 2008, 105, 3521–3526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, A.; Liu, S.V.; Subramaniam, D.S.; Giaccone, G. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat. Rev. Clin. Oncol. 2015, 12, 511–526. [Google Scholar] [CrossRef] [PubMed]
- Anasari, J.; Shackelford, R.E.; El-Osta, H. Epigenetics in non-small cell lung cancer: From basics to therapeutics. Transl. Lung Cancer Res. 2016, 5, 155–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ansari, J.; Yun, J.W.; Kompelli, A.R.; Moufarrej, Y.E.; Alexander, J.S.; Herrera, G.A.; Shackelford, R.E. The liquid biopsy in lung cancer. Genes Cancer 2016, 7, 355–367. [Google Scholar] [CrossRef] [Green Version]
- Jamal-Hanjani, M.; Wilson, G.A.; McGranahan, N.; Birkbak, N.J.; Watkins, T.B.K.; Veeriah, S.; Shafi, S.; Johnson, D.H.; Mitter, R.; Rosenthal, R.; et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 376, 2109–2121. [Google Scholar] [CrossRef] [Green Version]
- Mok, T.S.; Wu, Y.L.; Thongprasert, S.; Yang, C.H.; Chu, D.T.; Saijo, N.; Sunpaweravong, P.; Han, B.; Margono, B.; Ichinose, Y.; et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 2009, 361, 947–957. [Google Scholar] [CrossRef]
- Kim, E.S.; Hirsh, V.; Mok, T.; Socinski, M.A.; Gervais, R.; Wu, Y.L.; Li, L.Y.; Watkins, C.L.; Sellers, M.V.; Lowe, E.S.; et al. Gefitinib versus docetaxel in previously treated non-small-cell lung cancer (INTEREST): A randomized phase III trial. Lancet 2008, 372, 1809–1818. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Les acides nucléiques du plasma sanguin chez l’homme. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Chan, K.C.; Jiang, P.; Zheng, Y.W.; Liao, G.J.; Sun, H.; Wong, J.; Siu, S.S.; Chan, W.C.; Chan, S.L.; Chan, A.T.; et al. Cancer genome scanning in plasma: Detection of tumor-associated copy number aberrations; single-nucleotide variants; and tumoral heterogeneity by massively parallel sequencing. Clin. Chem. 2013, 59, 211–224. [Google Scholar] [CrossRef] [Green Version]
- Kotton, D.N.; Morrisey, E.E. Lung regeneration: Mechanisms; applications and emerging stem cell populations. Nat. Med. 2014, 20, 822–832. [Google Scholar] [CrossRef] [Green Version]
- Hanna, J.M.; Onaitis, M.W. Cell of origin of lung cancer. J. Carcinog. 2013, 12, 6. [Google Scholar] [CrossRef] [PubMed]
- Yeh, S.J.; Chang, C.A.; Li, C.W.; Wang, L.H.; Chen, B.S. Comparing progression molecular mechanisms between lung adenocarcinoma and lung squamous cell carcinoma based on genetic and epigenetic networks: Big data mining and genomewide systems identification. Oncotarget 2019, 10, 3760–3806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, K.S.; Liang, M.C.; Raiser, D.M.; Zamponi, R.; Roach, R.R.; Curtis, S.J. Characterization of the cell of origin for small cell lung cancer. Cell Cycle 2011, 10, 2806–2815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, K.; Tan, E.H.; O’Byrne, K.; Zhang, L.; Boyer, M.; Mok, T.; Hirsh, V.; Yang, J.C.; Lee, K.H.; Lu, S.; et al. Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation-positive non-small-cell lung cancer (LUX-Lung 7): A phase 2B; open-label; randomised controlled trial. Lancet Oncol. 2016, 17, 577–589. [Google Scholar] [CrossRef]
- Zhong, W.Z.; Wang, Q.; Mao, W.M.; Xu, S.T.; Wu, L.; Shen, Y.; Liu, Y.Y.; Chen, C.; Cheng, Y.; Xu, L.; et al. Gefitinib versus vinorelbine plus cisplatin as adjuvant treatment for stage II-IIIA (N1-N2) EGFR-mutant NSCLC (ADJUVANT/CTONG1104): A randomised; open-label; phase 3 study. Lancet Oncol. 2018, 19, 139–148. [Google Scholar] [CrossRef]
- Yoshino, I.; Osoegawa, A.; Yohena, T.; Kameyama, T.; Oki, E.; Oda, S.; Maehara, Y. Loss of heterozygosity (LOH) in non-small cell lung cancer: Difference between adenocarcinoma and squamous cell carcinoma. Respir. Med. 2005, 99, 308–312. [Google Scholar] [CrossRef] [Green Version]
- Fong, M.K.; Daniels, M.; Goh, F.; Yang, I.A.; Bowman, R. The Current and Future Role of Genomics; Bals, R., Ed.; Lung cancer. ERS Monogr: Gosport, UK, 2015; pp. 68–80. [Google Scholar]
- Gerber, D.E.; Paik, P.K.; Dowlati, A. Beyond adenocarcinoma: Current treatments and future directions for squamous; small cell; and rare lung cancer histologies. Am. Soc. Clin. Oncol. Educ. Book 2015, 147–162. [Google Scholar] [CrossRef]
- Shackelford, R.E.; Vora, M.; Mayhall, K.; Cotelingam, J. ALK-rearrangements and testing methods in non-small cell lung cancer: A review. Genes Cancer 2014, 5, 1–14. [Google Scholar] [CrossRef]
- Vanderlaan, P.A.; Yamaguchi, N.; Folch, E.; Boucher, D.H.; Kent, M.S.; Gangadharan, S.P.; Majid, A.; Goldstein, M.A.; Huberman, M.S.; Kocher, O.N.; et al. Success and failure rates of tumor genotyping techniques in routine pathological samples with non-small-cell lung cancer. Lung Cancer 2014, 84, 39–44. [Google Scholar] [CrossRef] [Green Version]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; Tarpey, P.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Ramnath, N.; Nagrath, S. Current Status of CTCs as Liquid Biopsy in Lung Cancer and Future Directions. Front. Oncol. 2015, 5, 209. [Google Scholar] [CrossRef] [PubMed]
- McAllister, S.S.; Weinberg, R.A. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat. Cell Biol. 2014, 16, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Q.; Stroun, M.; Magnenat, J.L.; Nicod, L.P.; Kurt, A.M.; Lyautey, J.; Lederrey, C.; Anker, P. Microsatellite alterations in plasma DNA of small cell lung cancer patients. Nat. Med. 1996, 2, 1033–1035. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Liu, D.; Shishodia, S.; Ozburn, N.; Behrens, C.; Lee, J.J.; Hong, W.K.; Aggarwal, B.B.; Wistuba, I.I. Nuclear factor-κB (NF-κB) is frequently expressed in lung cancer and preneoplastic lesions. Cancer 2006, 107, 2637–2646. [Google Scholar] [CrossRef]
- Saxon, J.A.; Yu, H.; Polosukhin, V.V.; Stathopoulos, G.T.; Gleaves, L.A.; McLoed, A.G.; Massion, P.P.; Yull, F.E.; Zhao, Z.; Blackwell, T.S. p52 expression enhances lung cancer progression. Sci. Rep. 2018, 8, 6078. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Gray, Z.; Willette-Brown, J.; Zhu, F.; Shi, G.; Jiang, Q.; Song, N.Y.; Dong, L.; Hu, Y. Macrophage inducible nitric oxide synthase circulates inflammation and promotes lung carcinogenesis. Cell Death Dis. 2018, 4, 46. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.X.; Rastetter, R.H.; Wilhelm, D. Noncoding RNAs: An Introduction. Adv. Exp. Med. Biol. 2016, 886, 13–32. [Google Scholar] [CrossRef]
- Brosnan, C.A.; Voinnet, O. The long and the short of noncoding RNAs. Curr. Opin. Cell Biol. 2009, 21, 416–425. [Google Scholar] [CrossRef]
- Braicu, C.; Zimta, A.A.; Harangus, A.; Iurca, I.; Irimie, A.; Coza, O.; Berindan-Neagoe, I. The Function of Noncoding RNAs in Lung Cancer Tumorigenesis. Cancers 2019, 11, 605. [Google Scholar] [CrossRef] [Green Version]
- Slack, F.J.; Weidhaas, J.B. MicroRNAs as a potential magic bullet in cancer. Future Oncol. 2006, 2, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Berindan-Neagoe, I.; Monroig, P.C.; Pasculli, B.; Calin, G.A. MicroRNAome Genome: A Treasure for Cancer Diagnosis and Therapy. CA Cancer J. Clin. 2014, 64, 311–336. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brambilla, E.; Gazdar, A. Pathogenesis of lung cancer signalling pathways: Roadmap for therapies. Eur. Respir. J. 2009, 33, 1485–1497. [Google Scholar] [CrossRef]
- Chan, L.W.; Wang, F.F.; Cho, W.C. Genomic sequence analysis of EGFR regulation by microRNAs in lung cancer. Curr. Top. Med. Chem. 2012, 12, 920–926. [Google Scholar] [CrossRef]
- Yan, C.; Zhang, W.; Shi, X.; Zheng, J.; Jin, X.; Huo, J. MiR-760 suppresses non-small cell lung cancer proliferation and metastasis by targeting ROS1. Environ. Sci. Pollut. Res. Int. 2018, 25, 18385–18391. [Google Scholar] [CrossRef]
- Vishwamitra, D.; Li, Y.; Wilson, D.; Manshouri, R.; Curry, C.V.; Shi, B.; Tang, X.M.; Sheehan, A.M.; Wistuba, I.I.; Shi, P.; et al. MicroRNA 96 is a post-transcriptional suppressor of anaplastic lymphoma kinase expression. Am. J. Pathol. 2012, 180, 1772–1780. [Google Scholar] [CrossRef] [Green Version]
- He, X.Y.; Chen, J.X.; Zhang, Z.; Li, C.L.; Peng, Q.L.; Peng, H.M. The let-7a microRNA protects from growth of lung carcinoma by suppression of k-Ras and c-Myc in nude mice. J. Cancer Res. Clin. Oncol. 2010, 136, 1023–1028. [Google Scholar] [CrossRef]
- Seviour, E.G.; Sehgal, V.; Mishra, D.; Rupaimoole, R.; Rodriguez-Aguayo, C.; Lopez-Berestein, G.; Lee, J.S.; Sood, A.K.; Kim, M.P.; Mills, G.B.; et al. Targeting KRas-dependent tumour growth; circulating tumour cells and metastasis in vivo by clinically significant miR-193a-3p. Oncogene 2017, 36, 1339–135031. [Google Scholar] [CrossRef] [Green Version]
- Qiu, X.; Wang, D.; Li, Y.; Zhang, B.; Li, Y.; Zhang, B.; Yuan, T.; Wei, J.; Zhao, B.; Zhao, X.; et al. MiR-181a-5p inhibits cell proliferation and migration by targeting Kras in non-small cell lung cancer A549 cells. Acta Biochim. Biophys. Sin. 2015, 47, 630–638. [Google Scholar] [CrossRef] [Green Version]
- Xie, Q.; Yu, Z.; Lu, Y.; Fan, J.; Ni, Y.; Ma, L. microRNA-148a-3p inhibited the proliferation and epithelial-mesenchymal transition progression of non-small-cell lung cancer via modulating Ras/MAPK/Erk signaling. J. Cell. Physiol. 2019, 234, 12786–12799. [Google Scholar] [CrossRef]
- Jiang, W.; Wei, K.; Pan, C.; Li, H.; Cao, J.; Han, X.; Tang, Y.; Zhu, S.; Yuan, W.; He, Y.; et al. MicroRNA-1258 suppresses tumour progression via GRB2/Ras/Erk pathway in non-small-cell lung cancer. Cell Prolif. 2018, 51, e12502. [Google Scholar] [CrossRef]
- Lv, X.; Li, C.Y.; Han, P.; Xu, X.Y. MicroRNA-520a-3p inhibits cell growth and metastasis of non-small cell lung cancer through PI3K/AKT/mTOR signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2321–2327. [Google Scholar] [CrossRef] [PubMed]
- Fortunato, O.; Boeri, M.; Moro, M.; Verri, C.; Mensah, M.; Conte, D.; Caleca, L.; Roz, L.; Pastorino, U.; Sozzi, G. Mir-660 is downregulated in lung cancer patients and its replacement inhibits lung tumorigenesis by targeting MDM2-p53 interaction. Cell Death Dis. 2014, 5, e1564. [Google Scholar] [CrossRef] [Green Version]
- Kong, Q.; Shu, N.; Li, J.; Xu, N. miR-641 Functions as a Tumor Suppressor by Targeting MDM2 in Human Lung Cancer. Oncol. Res. 2018, 26, 735–741. [Google Scholar] [CrossRef]
- Bandi, N.; Zbinden, S.; Gugger, M.; Arnold, M.; Kocher, V.; Hasan, L.; Kappeler, A.; Brunner, T.; Vassella, E. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009, 69, 5553–5559. [Google Scholar] [CrossRef] [Green Version]
- Ren, X.S.; Yin, M.H.; Zhang, X.; Wang, Z.; Feng, S.P.; Wang, G.X.; Luo, Y.J.; Liang, P.Z.; Yang, X.Q.; He, J.X.; et al. Tumor-suppressive microRNA-449a induces growth arrest and senescence by targeting E2F3 in human lung cancer cells. Cancer Lett. 2014, 344, 195–203. [Google Scholar] [CrossRef]
- Chen, L.; Gibbons, D.L.; Goswami, S.; Cortez, M.A.; Ahn, Y.H.; Byers, L.A.; Zhang, X.; Yi, X.; Dwyer, D.; Lin, W.; et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 2014, 5, 5241. [Google Scholar] [CrossRef]
- Pecot, C.V.; Rupaimoole, R.; Yang, D.; Yang, D.; Akbani, R.; Ivan, C.; Wu, S.; Han, H.D.; Shah, M.Y.; Rodriguez-Aguayo, C.; et al. Tumour angiogenesis regulation by the miR-200 family. Nat. Commun. 2013, 4, 2427. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Peng, X.C.; Zheng, X.L.; Wang, J.; Qin, Y.W. MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer 2009, 66, 169–175. [Google Scholar] [CrossRef]
- Hu, J.; Cheng, Y.; Li, Y.; Jin, Z.; Pan, Y.; Liu, G.; Fu, S.; Zhang, Y.; Feng, K.; Feng, Y. microRNA-128 plays a critical role in human non-small cell lung cancer tumourigenesis; angiogenesis and lymphangiogenesis by directly targeting vascular endothelial growth factor-C. Eur. J. Cancer 2014, 50, 2336–2350. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Liu, Y.; Fang, X.; Liu, Y.; Fang, L.; Lin, L.; Liu, X.; Wang, N. Tumor-derived microRNA-494 promotes angiogenesis in non-small cell lung cancer. Angiogenesis 2015, 18, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.Y.; Wang, Y.; An, Z.J.; Shi, C.G.; Zhu, G.A.; Wang, B.; Lu, M.Y.; Pan, C.K.; Chen, P. Downregulation of miR-497 promotes tumor growth and angiogenesis by targeting HDGF in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2013, 435, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Gao, J.; Huang, Q.; Jin, Y.; Wei, Z. Downregulating microRNA-144 mediates a metabolic shift in lung cancer cells by regulating GLUT1 expression. Oncol. Lett. 2016, 11, 3772–3776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, G.; Huang, G.; Liu, H.D.; Liang, H.X.; Ni, Y.F.; Ding, Z.H.; Ni, G.Y.; Hua, H.W. MiR-199a suppresses the hypoxia-induced proliferation of non-small cell lung cancer cells through targeting HIF1alpha. Mol. Cell. Biochem. 2013, 384, 173–180. [Google Scholar] [CrossRef]
- Zhai, S.; Zhao, L.; Lin, T.; Wang, W. Downregulation of miR-33b promotes non-small cell lung cancer cell growth through reprogramming glucose metabolism miR-33b regulates non-small cell lung cancer cell growth. J. Cell. Biochem. 2019, 120, 6651–6660. [Google Scholar] [CrossRef] [Green Version]
- Roth, A.; Diederichs, S. Long Noncoding RNAs in Lung Cancer. Curr. Top. Microbiol. Immunol. 2016, 394, 57–110. [Google Scholar] [CrossRef]
- Nakagawa, T.; Endo, H.; Yokoyama, M.; Abe, J.; Tamai, K.; Tanaka, N.; Sato, I.; Takahashi, S.; Kondo, T.; Satoh, K. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2013, 436, 319–324. [Google Scholar] [CrossRef]
- Cortez, M.A.; Ivan, C.; Valdecanas, D.; Wang, X.; Peltier, H.J.; Ye, Y.; Araujo, L.; Carbone, D.P.; Shilo, K.; Giri, D.K.; et al. PDL1 Regulation by p53 via miR-34. J. Natl. Cancer Inst. 2015, 108, djv303. [Google Scholar] [CrossRef] [Green Version]
- Fujita, Y.; Yagishita, S.; Hagiwara, K.; Yoshioka, Y.; Kosaka, N.; Takeshita, F.; Fujiwara, T.; Tsuta, K.; Nokihara, H.; Tamura, T.; et al. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol. Ther. 2015, 23, 717–727. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Chen, J.; Dai, J.; Zhang, B.; Wang, F.; Sun, Y. MicroRNA-16-1 Inhibits Tumor Cell Proliferation and Induces Apoptosis in A549 Non-Small Cell Lung Carcinoma Cells. Oncol. Res. 2016, 24, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Hu, L.; Li, X.; Geng, J.; Dai, M.; Bai, X. MicroRNA-130b promotes lung cancer progression via PPARgamma/VEGF-A/BCL-2-mediated suppression of apoptosis. J. Exp. Clin. Cancer Res. 2016, 35, 105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Liu, C.; Niu, Y.; Jing, Y.; Zhang, H.; Wang, J.; Yang, J.; Zen, K.; Zhang, J.; Zhang, C.Y.; et al. MicroRNA-128-3p regulates mitomycin C-induced DNA damage response in lung cancer cells through repressing SPTAN1. Oncotarget 2017, 8, 58098–58107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vincent, K.; Pichler, M.; Lee, G.W.; Ling, H. MicroRNAs, genomic instability and cancer. Int. J. Mol. Sci. 2014, 15, 14475–14491. [Google Scholar] [CrossRef] [Green Version]
- Zhu, B.; Cao, X.; Zhang, W.; Pan, G.; Yi, Q.; Zhong, W.; Yan, D. MicroRNA-31-5p enhances the Warburg effect via targeting FIH. FASEB J. 2018, 33, 545–556. [Google Scholar] [CrossRef] [Green Version]
- Guo, T.; Li, J.; Zhang, L.; Hou, W.; Wang, R.; Zhang, J.; Gao, P. Multidimensional communication of microRNAs and long noncoding RNAs in lung cancer. J. Cancer Res. Clin. Oncol. 2019, 145, 31–48. [Google Scholar] [CrossRef]
- Gutschner, T.; Diederichs, S. The hallmarks of cancer: A long noncoding RNA point of view. RNA Biol. 2012, 9, 703–719. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Yu, M.; Chang, Q. Mdig de-represses H19 large intergenic noncoding RNA (lincRNA) by down-regulating H3K9me3 and heterochromatin. Oncotarget 2013, 4, 1427–1437. [Google Scholar] [CrossRef] [Green Version]
- Matouk, I.J.; Mezan, S.; Mizrahi, A.; Ohana, P.; Abu-Lail, R.; Fellig, Y.; Degroot, N.; Galun, E.; Hochberg, A. The oncofetal H19 RNA connection: Hypoxia; p53 and cancer. Biochim. Biophys. Acta 2010, 1803, 443–451. [Google Scholar] [CrossRef] [Green Version]
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1; a novel noncoding RNA; and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, L.H.; Spieker, T.; Koschmieder, S.; Schäffers, S.; Humberg, J.; Jungen, D.; Bulk, E.; Hascher, A.; Wittmer, D.; Marra, A.; et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2011, 6, 1984–1992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Li, X.; Li, X.; Chen, Z. LncRNA H19 promotes epithelial-mesenchymal transition (EMT) by targeting miR-484 in human lung cancer cells. J. Cell Biochem. 2018, 119, 4447–4457. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.J.; Lin, Q.; Ma, J.X.; Shi, W.W.; Yang, B.; Li, F. Long noncoding RNA NEAT1 acts as oncogene in NSCLC by regulating the Wnt signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 504–510. [Google Scholar] [PubMed]
- Wang, P.; Chen, D.; Ma, H.; Li, Y. LncRNA SNHG12 contributes to multidrug resistance through activating the MAPK/Slug pathway by sponging miR-181a in non-small cell lung cancer. Oncotarget 2017, 8, 84086–84101. [Google Scholar] [CrossRef] [Green Version]
- Cao, L.; Chen, J.; Ou, B.; Liu, C.; Zou, Y.; Chen, Q. GAS5 knockdown reduces the chemo-sensitivity of non-small cell lung cancer (NSCLC) cell to cisplatin (DDP) through regulating miR-21/ PTEN axis. Biomed. Pharmacother. 2017, 93, 570–579. [Google Scholar] [CrossRef]
- Tang, Y.; He, R.; An, J.; Deng, P.; Huang, L.; Yang, W. lncRNA XIST interacts with miR-140 to modulate lung cancer growth by targeting iASPP. Oncol. Rep. 2017, 38, 941–948. [Google Scholar] [CrossRef] [Green Version]
- Mitsudomi, T.; Morita, S.; Yatabe, Y.; Negoro, S.; Okamoto, I.; Tsurutani, J.; Seto, T.; Satouchi, M.; Tada, H.; Hirashima, T.; et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label; randomised phase 3 trial. Lancet Oncol. 2010, 11, 121–128. [Google Scholar] [CrossRef]
- Shaw, A.T.; Kim, D.W.; Nakagawa, K.; Seto, T.; Crinó, L.; Ahn, M.J.; De Pas, T.; Besse, B.; Solomon, B.J.; Blackhall, F.; et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N. Engl. J. Med. 2013, 368, 2385–2394. [Google Scholar] [CrossRef] [Green Version]
- Taneja, T.K.; Sharma, S. Markers of small cell lung cancer. World J. Surg. Oncol. 2004, 2, 10. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.S.; Herbst, R.S.; Wistuba, I.I.; Lee, J.J.; Blumenschein, G.R., Jr.; Tsao, A.; Stewart, D.J.; Hicks, M.E.; Erasmus, J., Jr.; Gupta, S.; et al. The BATTLE trial: Personalizing therapy for lung cancer. Cancer Discov. 2011, 1, 44–53. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.Y.; Zhong, W.Z.; Zhang, X.C.; Su, J.; Yang, X.N.; Chen, Z.H.; Yang, J.J.; Zhou, Q.; Yan, H.H.; An, S.J.; et al. EGFR mutation heterogeneity and the mixed response to EGFR tyrosine kinase inhibitors of lung adenocarcinomas. Oncologist 2012, 17, 978–985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sargent, D.J.; Conley, B.A.; Allegra, C.; Collette, L. Clinical trial designs for predictive marker validation in cancer treatment trials. J. Clin. Oncol. 2005, 23, 2020–2027. [Google Scholar] [CrossRef] [PubMed]
- Freidlin, B.; McShane, L.M.; Polley, M.Y.; Korn, E.L. Randomized phase II trial designs with biomarkers. J. Clin. Oncol. 2012, 30, 3304–3309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018, 36, 1631–1641. [Google Scholar] [CrossRef] [PubMed]
- Oxnard, G.R.; Paweletz, C.P.; Sholl, L.M. Genomic analysis of plasma cell-free DNA in patients with cancer. JAMA Oncol. 2017, 3, 740–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thress, K.S.; Brant, R.; Carr, T.H.; Dearden, S.; Jenkins, S.; Brown, H.; Hammett, T.; Cantarini, M.; Barrett, J.C. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015, 90, 509–515. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.; Kang, X.; You, X.; Dai, L.; Tian, D.; Yan, W.; Yang, Y.; Xiong, H.; Liang, Z.; Zhao, G.Q.; et al. Cross-platform comparison of four leading technologies for detecting EGFR mutations in circulating tumor DNA from non-small cell lung carcinoma patient plasma. Theranostics 2017, 7, 1437–1446. [Google Scholar] [CrossRef]
- Mao, X.; Zhang, Y.; Xie, F.; Zheng, X.; Sun, J. Can peripheral blood be used as surrogate in detecting epidermal growth factor receptor mutation status in advanced non-small cell lung cancer patients? A meta-analysis. Oncotarget 2017, 8, 78057–78067. [Google Scholar] [CrossRef] [Green Version]
- Madsen, A.T.; Hojbjerg, J.A.; Sorensen, B.S.; Winther-Larsen, A. Day-to-day and within-day biological variation of cell-free DNA. EBioMedicine 2019, 49, 284–290. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.H.; Montalvo, L.; Chrebtow, V.; Busch, M.P. Quantitation of genomic DNA in plasma and serum samples: Higher concentrations of genomic DNA found in serum than in plasma. Transfusion 2001, 41, 276–282. [Google Scholar] [CrossRef]
- Huang, W.L.; Chen, Y.L.; Yang, S.C.; Ho, C.L.; Wei, F.; Wong, D.T.; Su, W.C.; Lin, C.C. Liquid biopsy genotyping in lung cancer: Ready for clinical utility? Oncotarget 2017, 8, 18590–18608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa- Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vykoukal, J.; Sun, N.; Aguilar-Bonavides, C.; Katayama, H.; Tanaka, I.; Fahrmann, J.F.; Capello, M.; Fujimoto, J.; Aguilar, M.; Wistuba, I.I.; et al. Plasma-derived extracellular vesicle proteins as a source of biomarkers for lung adenocarcinoma. Oncotarget 2017, 8, 95466–95480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krug, A.K.; Enderle, D.; Karlovich, C.; Priewasser, T.; Bentink, S.; Spiel, A.; Brinkmann, K.; Emenegger, J.; Grimm, D.G.; Castellanos-Rizaldos, E.; et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann. Oncol. 2018, 29, 700–706. [Google Scholar] [CrossRef] [Green Version]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 3, 255–289. [Google Scholar] [CrossRef]
- Castellanos-Rizaldos, E.; Grimm, D.G.; Tadigotla, V.; Hurley, J.; Healy, J.; Neal, P.L.; Sher, M.; Venkatesan, R.; Karlovich, C.; Raponi, M.; et al. Exosome-based Detection of EGFR T790M in Plasma from Non-Small Cell Lung Cancer Patients. Clin Cancer Res. 2018, 24, 2944–2950. [Google Scholar] [CrossRef] [Green Version]
- Okamura, K.; Takayama, K.; Izumi, M.; Harada, T.; Furuyama, K.; Nakanishi, Y. Diagnostic value of CEA and CYFRA 21-1 tumor markers in primary lung cancer. Lung Cancer 2013, 80, 45–49. [Google Scholar] [CrossRef]
- Huang, L.; Zhou, J.G.; Yao, W.X.; Tian, X.; Lv, S.P.; Zhang, T.Y.; Jin, S.H.; Bai, Y.J.; Ma, H. Systematic review and meta-analysis of the efficacy of serum neuron-specific enolase for early small cell lung cancer screening. Oncotarget 2017, 8, 64358–64372. [Google Scholar] [CrossRef] [Green Version]
- Mok, T.; Wu, Y.L.; Lee, J.S.; Yu, C.J.; Sriuranpong, V.; Sandoval-Tan, J.; Ladrera, G.; Thongprasert, S.; Srimuninnimit, V.; Liao, M.; et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res. 2015, 21, 3196–3203. [Google Scholar] [CrossRef] [Green Version]
- Sacher, A.G.; Paweletz, C.; Dahlberg, S.E.; Alden, R.S.; O’Connell, A.; Feeney, N.; Mach, S.L.; Jänne, P.A.; Oxnard, G.R. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016, 2, 1014–1022. [Google Scholar] [CrossRef] [Green Version]
- Marchetti, A.; Palma, J.F.; Felicioni, L.; De Pas, T.M.; Chiari, R.; Del Grammastro, M.; Filice, G.; Ludovini, V.; Brandes, A.A.; Chella, A.; et al. Early prediction of response to tyrosine kinase inhibitors by quantification of EGFR mutations in plasma of NSCLC patients. J. Thorac. Oncol. 2015, 10, 1437–1443. [Google Scholar] [CrossRef] [Green Version]
- Sorensen, B.S.; Wu, L.; Wei, W.; Tsai, J.; Weber, B.; Nexo, E.; Meldgaard, P. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer 2014, 120, 3896–3901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Boeri, M.; Pastorino, U.; Sozzi, G. Role of microRNAs in lung cancer: microRNA signatures in cancer prognosis. Cancer J. 2012, 18, 268–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mensah, M.; Borzi, C.; Verri, C.; Suatoni, P.; Conte, D.; Pastorino, U.; Orazio, F.; Sozzi, G.; Boeri, M. MicroRNA Based Liquid Biopsy: The Experience of the Plasma miRNA Signature Classifier (MSC) for Lung Cancer Screening. J. Vis. Exp. 2017, 128. [Google Scholar] [CrossRef]
- Moretti, F.; D’Antona, P.; Finardi, E.; Barbetta, M.; Lorenzo, D.; Poli, A.; Gini, E.; Noonan, D.M.; Imperatori, A.; Rotolo, N.; et al. Systematic review and critique of circulating miRNAs as biomarkers of stage I-II non-small cell lung cancer. Oncotarget 2017, 8, 94980–94996. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Hu, Z.; Zhou, Y.; Zhao, G.; Lei, Y.; Li, G.; Chen, S.; Chen, K.; Shen, Z.; Chen, X.; et al. The clinical use of circulating microRNAs as noninvasive diagnostic biomarkers for lung cancers. Oncotarget 2017, 8, 90197–90214. [Google Scholar] [CrossRef] [Green Version]
- Qu, L.; Li, L.; Zheng, X.; Fu, H.; Tang, C.; Qin, H.; Li, X.; Wang, H.; Li, J.; Wang, W.; et al. Circulating plasma microRNAs as potential markers to identify EGFR mutation status and to monitor epidermal growth factor receptor-tyrosine kinase inhibitor treatment in patients with advanced non-small cell lung cancer. Oncotarget 2017, 8, 45807–45824. [Google Scholar] [CrossRef] [Green Version]
- D’Antona, P.; Cattoni, M.; Dominioni, L.; Poli, A.; Moretti, F. Serum miR-223: A Validated Biomarker for Detection of Early-Stage Non-Small Cell Lung Cancer. Cancer Epidemiol. Biomark. 2019, 28, 1926–1933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.L.; Qu, L.L.; Fu, H.J.; Zheng, X.F.; Tang, C.H.; Li, X.Y.; Chen, J.; Wang, W.X.; Yang, S.X.; Wang, L.; et al. Circulating microRNAs as novel biomarkers of ALK-positive nonsmall cell lung cancer and predictors of response to crizotinib therapy. Oncotarget 2017, 8, 45399–45414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, D.G.; Johnen, G.; Casjens, S.; Bryk, O.; Pesch, B.; Jöckel, K.H.; Kollmeier, J.; Brüning, T. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res. Notes 2013, 6, 518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, F.; Yu, F.; Wang, J.; Li, Y.; Li, Y.; Li, Z.; Zhou, Q. Expression of MALAT1 in the peripheral whole blood of patients with lung cancer. Biomed. Rep. 2015, 3, 309–312. [Google Scholar] [CrossRef] [Green Version]
- Thai, P.; Statt, S.; Chen, C.H.; Liang, E.; Campbell, C.; Wu, R. Characterization of a novel long noncoding RNA.; SCAL1; induced by cigarette smoke and elevated in lung cancer cell lines. Am. J. Respir. Cell Mol. Biol. 2013, 49, 204–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tantai, J.; Hu, D.; Yang, Y.; Geng, J. Combined identification of long noncoding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 7887–7895. [Google Scholar]
- Xie, Y.; Zhang, Y.; Du, L.; Jiang, X.; Yan, S.; Duan, W.; Li, J.; Zhan, Y.; Wang, L.; Zhang, S.; et al. Circulating long noncoding RNA act as potential novel biomarkers for diagnosis and prognosis of non-small cell lung cancer. Mol. Oncol. 2018, 12, 648–658. [Google Scholar] [CrossRef]
- Hu, X.; Bao, J.; Wang, Z.; Zhang, Z.; Gu, P.; Tao, F.; Cui, D.; Jiang, W. The plasma lncRNA acting as fingerprint in nonsmall-cell lung cancer. Tumour Biol. 2016, 37, 3497–3504. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, L.; Yan, S.; Li, W.; Cui, J.; Zhu, M.; Xia, N.; Yang, Y.; Yuan, J.; Chen, X.; et al. LncRNA16 is a potential biomarker for diagnosis of early-stage lung cancer that promotes cell proliferation by regulating the cell cycle. Oncotarget 2017, 8, 7867–7877. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.; Lv, T.; Shi, X.; Liu, H.; Zhu, Q.; Zeng, J.; Yang, W.; Yin, J.; Song, Y. Circulating long noncoding RNA GAS5 is a novel biomarker for the diagnosis of nonsmall cell lung cancer. Medicine (Baltimore) 2016, 95, e4608. [Google Scholar] [CrossRef]
- Jenkins, S.; Yang, J.C.; Ramalingam, S.S.; Yu, K.; Patel, S.; Weston, S.; Hodge, R.; Cantarini, M.; Jänne, P.A.; Mitsudomi, T.; et al. Plasma ctDNA analysis for detection of the EGFR T790M mutation in patients with advanced non-small cell lung cancer. J. Thorac. Oncol. 2017, 12, 1061–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, X.; Liu, J.; Sun, Y.; Wang, M.; Lei, H.; Luo, G.; Liu, X.; Xiong, C.; Liu, D.; Liu, J.; et al. Circulating cell-free DNA has a high degree of specificity to detect exon 19 deletions and the single-point substitution mutation L858R in non-small cell lung cancer. Oncotarget 2016, 7, 29154–29165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, M.; Wang, J.; Xu, Y.; Ding, X.; Li, M.; Jiang, F.; Xu, L.; Yin, R. Circulating tumor DNA is effective for the detection of EGFR mutation in non-small cell lung cancer: A meta-analysis. Cancer Epidemiol. Biomark. Prev. 2015, 24, 206–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, J.; Shen, L.; Zheng, D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: A systematic review and meta-analysis. Sci. Rep. 2014, 4, 6269. [Google Scholar] [CrossRef]
- Han, B.; Tjulandin, S.; Hagiwara, K.; Normanno, N.; Wulandari, L.; Laktionov, K.; Hudoyo, A.; He, Y.; Zhang, Y.P.; Wang, M.Z.; et al. EGFR mutation prevalence in Asia-Pacific and Russian patients with advanced NSCLC of adenocarcinoma and non-adenocarcinoma histology: The IGNITE study. Lung Cancer 2017, 113, 37–44. [Google Scholar] [CrossRef]
- Reck, M.; Hagiwara, K.; Han, B.; Tjulandin, S.; Grohe, C.; Yokoi, T.; Morabito, A.; Novello, S.; Arriola, E.; Molinier, O.; et al. ctDNA Determination of EGFR Mutation Status in European and Japanese Patients with Advanced NSCLC: The ASSESS Study. J. Thorac. Oncol. 2016, 11, 1682–1689. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.A.; Arcila, M.E.; Rekhtman, N.; Sima, C.S.; Zakowski, M.F.; Pao, W.; Kris, M.G.; Miller, V.A.; Ladanyi, M.; Riely, G.J. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin. Cancer Res. 2013, 19, 2240–2247. [Google Scholar] [CrossRef] [Green Version]
- Tan, C.S.; Cho, B.C.; Soo, R.A. Next-generation epidermal growth factor receptor tyrosine kinase inhibitors in epidermal growth factor receptor -mutant non-small cell lung cancer. Lung Cancer 2016, 93, 59–68. [Google Scholar] [CrossRef]
- Oxnard, G.R.; Thress, K.S.; Alden, R.S.; Lawrance, R.; Paweletz, C.P.; Cantarini, M.; Yang, J.C.; Barrett, J.C.; Jänne, P.A. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non- Small-Cell Lung Cancer. J. Clin. Oncol. 2016, 34, 3375–3382. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Chen, R.; Wang, S.; Zhong, J.; Wu, M.; Zhao, J.; Duan, J.; Zhuo, M.; An, T.; Wang, Y.; et al. Quantification and dynamic monitoring of EGFR T790M in plasma cell-free DNA by digital PCR for prognosis of EGFR-TKI treatment in advanced NSCLC. PLoS ONE 2019, 9, e110780. [Google Scholar] [CrossRef]
- Oxnard, G.R.; Thress, K.S.; Alden, R.S.; Lawrance, R.; Paweletz, C.P.; Cantarini, M.; Barrett, C.; Yang, J.; Jänne, P.A. 135O_PR: Plasma genotyping for predicting benefit from osimertinib in patients (pts) with advanced NSCLC. J. Thorac. Oncol. 2016, 11 (Suppl. 4), S154. [Google Scholar] [CrossRef]
- Rolfo, C.; Laes, J.F.; Reclusa, P.; Valentino, A.; Lienard, M.; Gil-Bazo, I.; Malapelle, U.; Sirera, R.; Rocco, D.; Meerbeeck, V.J.; et al. P2.01-093 Exo-ALK Proof of Concept: Exosomal Analysis of ALK Alterations in Advanced NSCLC Patients. J. Thorac. Oncol. 2017, 12, S844–S845. [Google Scholar] [CrossRef] [Green Version]
- Ilié, M.; Szafer-Glusman, E.; Hofman, V.; Chamorey, E.; Lalvée, S.; Selva, E.; Leroy, S.; Marquette, C.H.; Kowanetz, M.; Hedge, P.; et al. Detection of PD-L1 in circulating tumor cells and white blood cells from patients with advanced non-small-cell lung cancer. Ann. Oncol. 2018, 29, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Szafer-Glusman, E.; Hofman, V.; Long-Mira, E.; Suttmann, R.; Darbonne, W.; Butori, C.; Lalvée, S.; Fayada, J.; Selva, E.; et al. Expression of MET in circulating tumor cells correlates with expression in tumor tissue from advanced-stage lung cancer patients. Oncotarget 2017, 8, 26112–26121. [Google Scholar] [CrossRef] [Green Version]
- Remon, J.; Caramella, C.; Jovelet, C.; Lacroix, L.; Lawson, A.; Smalley, S.; Howarth, K.; Gale, D.; Green, E.; Plagnol, V.; et al. Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann. Oncol. 2017, 28, 784–790. [Google Scholar] [CrossRef]



| Histological Type of Lung Cancer | Tumor Suppressor Genes | Oncogenes | References |
|---|---|---|---|
| Adenocarcinoma | TP53 Mt 90% 17p LOH 70% 9p LOH 50–75% 3p allele loss 50–80% 13q LOH 40–60% CDKN2A 10–40% | KRAS Mt 32.2% EGFR Mt 15% FGFR4 Mt 7% EML4/ALK Fus 1–5% BRAF Mt 2–3% MET Mt/Amp 1–20% Her2 Mt 0.9% | [12,24,25,26,27,28] |
| Squamous cell carcinoma | FGFR1 Amp 20% PIK3CA Mt 12% KRAS Mt 6% EGFR Mt 5% BRAF Mt 2% | [18,24,25,26,29,30,31] | |
| SCLC | 17p LOH 80–90% 13q LOH 75% 9p LOH 20–50% 3p allele loss >90% TP53 Mt 90% Absent RB1 expression 90% | KRAS Mt 25% RET Fus 5% EGFR Mt 4% ROS1 Fus 1% PIK3CA Mt 3% MYC Amp 15–30% | [25,32,33,34] |
| Hallmark of Cancer | Target of miRNA | miRNA | Reference |
|---|---|---|---|
| Sustaining proliferative signaling | EGFR ROS1 ALK KRAS PI3K | miR-7, miR-27a-3p, miR-30, miR-34, miR-128, miR-133, miR-134, miR-145, miR-146, miR-149, miR-218, miR-542-5p, miR-760, miR-96, let-7, miR-193a-3p, miR-181a-5p, miR-148a-3p (via SOS2), miR-1258 (via GRB2), miR-520a-3p | [45,46,47,48,49,50,51,52,53,54] |
| Evading growth suppression | MDM2/p53 RB E2F | miR-641, miR-660, miR-15/miR-16, miR-449a | [55,56,57,58] |
| Enabling replicative immortality | No miRNAs reported to target hTERT in lung cancer studies | ||
| Invasion and metastasis | ZEB1 and ZEB2 | miR-200 | [59] |
| Inducing angiogenesis | VEGF PTEN | mir-200, miR-126, miR128, miR-494, miR-497 | [60,61,62,63,64] |
| Deregulating cellular energetics | GLUT1 HIF-1α LDHA | miR-144, miR-199a, miR-31-5p, miR-33b | [65,66,67,68,69] |
| Avoiding immune destruction and tumor-promoting inflammation | PD-L1 | miR-34a, miR-197, miR-200 family | [59,70,71] |
| Resisting cell death | Bcl-2 | miR-16-1, miR-130b (via PPAR-γ/VEGF) | [72,73] |
| Genome instability and mutation | SPTAN1 P53/PIK3R1 and CDC42 | miR-128-3p, miR-125b, miR-504, miR-29 | [74,75] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haranguș, A.; Berindan-Neagoe, I.; Todea, D.A.; Șimon, I.; Șimon, M. Noncoding RNAs and Liquid Biopsy in Lung Cancer: A Literature Review. Diagnostics 2019, 9, 216. https://doi.org/10.3390/diagnostics9040216
Haranguș A, Berindan-Neagoe I, Todea DA, Șimon I, Șimon M. Noncoding RNAs and Liquid Biopsy in Lung Cancer: A Literature Review. Diagnostics. 2019; 9(4):216. https://doi.org/10.3390/diagnostics9040216
Chicago/Turabian StyleHaranguș, Antonia, Ioana Berindan-Neagoe, Doina Adina Todea, Ioan Șimon, and Mărioara Șimon. 2019. "Noncoding RNAs and Liquid Biopsy in Lung Cancer: A Literature Review" Diagnostics 9, no. 4: 216. https://doi.org/10.3390/diagnostics9040216