Direct Detection of Glycated Human Serum Albumin and Hyperglycosylated IgG3 in Serum, by MALDI-ToF Mass Spectrometry, as a Predictor of COVID-19 Severity
Abstract
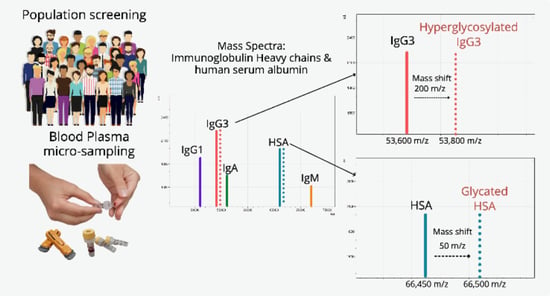
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Sample Analysis by MALDI-ToF Mass Spectrometry
2.3. Spectral Data Processing
2.4. Statistical Analysis
3. Results
3.1. Direct Plasma Total IgG1 Spectra
3.2. Direct Plasma Total IgG3 Spectra
3.3. Direct Plasma HSA Spectra
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| AGE | Advanced glycation end products |
| ARDS | Acute respiratory distress syndrome |
| AU | Arbitrary units |
| ECLIA | Electro-chemiluminescence assay |
| GDP | Gross domestic product |
| Hc | Heavy chain |
| HCWs | Health care workers, |
| HSA | Human serum albumin |
| Ig | Immunoglobulin |
| MALDI-ToF MS | Matrix assisted laser desorption ionization—time of flight. |
| ML | Machine learning |
| PCR | Polymerase chain reaction |
| RBD | Receptor binding domain |
| SA | Sinapinic acid |
| TCEP | Tris(2-carboxyethyl)phosphine |
References
- Iacobucci, G. COVID-19: Universal screening is likely to miss infected people, review finds. BMJ 2020, 370, m3619. [Google Scholar] [CrossRef] [PubMed]
- García-Montero, C.; Fraile-Martínez, O.; Bravo, C.; Torres-Carranza, D.; Sanchez-Trujillo, L.; Gómez-Lahoz, A.M.; Guijarro, L.G.; García-Honduvilla, N.; Asúnsolo, A.; Bujan, J.; et al. An Updated Review of SARS-CoV-2 Vaccines and the Importance of Effective Vaccination Programs in Pandemic Times. Vaccines 2021, 9, 433. [Google Scholar] [CrossRef] [PubMed]
- Rubin, R. COVID-19 Vaccines vs Variants—Determining How Much Immunity Is Enough. JAMA 2021, 325, 1241–1243. [Google Scholar] [CrossRef] [PubMed]
- Sobotka, T.; Brzozowska, Z.; Muttarak, R.; Zeman, K.; di Lego, V. Age, gender and COVID-19 infections. medRxiv 2020. [Google Scholar] [CrossRef]
- Mahase, E. COVID-19: Death rate is 0.66% and increases with age, study estimates. BMJ Br. Med. J. 2020, 369, m1327. [Google Scholar] [CrossRef] [Green Version]
- Han, E.; Tan, M.; Turk, E.; Sridhar, D.; Leung, G.M.; Shibuya, K.; Asgari, N.; Oh, J.; García-Basteiro, A.L.; Hanefeld, J.; et al. Lessons learnt from easing COVID-19 restrictions: An analysis of countries and regions in Asia Pacific and Europe. Lancet 2020, 396, 1525–1534. [Google Scholar] [CrossRef]
- Booth, A.; Reed, A.B.; Ponzo, S.; Yassaee, A.; Aral, M.; Plans, D.; Labrique, A.; Mohan, D. Population risk factors for severe disease and mortality in COVID-19: A global systematic review and meta-analysis. PLoS ONE 2021, 16, e0247461. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Hasegawa, K.; Ma, B.; Fujiogi, M.; Camargo, C.A.; Liang, L. Association of obesity and its genetic predisposition with the risk of severe COVID-19: Analysis of population-based cohort data. Metabolism 2020, 112, 154345. [Google Scholar] [CrossRef] [PubMed]
- Iles, R.K.; Makhzoumi, T.S. Explain Again: Why Are We Vaccinating Young Children against COVID-19? COVID 2022, 2, 492–500. [Google Scholar] [CrossRef]
- Clift, A.K.; Coupland, C.A.C.; Keogh, R.H.; Diaz-Ordaz, K.; Williamson, E.; Harrison, E.M.; Hayward, A.; Hemingway, H.; Horby, P.; Mehta, N.; et al. Living risk prediction algorithm (QCOVID) for risk of hospital admission and mortality from coronavirus 19 in adults: National derivation and validation cohort study. BMJ 2020, 371, m3731. [Google Scholar] [CrossRef]
- Wise, J. COVID-19: Extra 1.7 million people in England are asked to shield. BMJ 2021, 372, n467. [Google Scholar] [CrossRef]
- Cave, S.; Whittlestone, J.; Nyrup, R.; O hEigeartaigh, S.; Calvo, R.A. Using AI ethically to tackle COVID-19. BMJ 2021, 372, n364. [Google Scholar] [CrossRef] [PubMed]
- Assaf, D.; Gutman, Y.; Neuman, Y.; Segal, G.; Amit, S.; Gefen-Halevi, S.; Shilo, N.; Epstein, A.; Mor-Cohen, R.; Biber, A.; et al. Utilization of machine-learning models to accurately predict the risk for critical COVID-19. Intern. Emerg. Med. 2020, 15, 1435–1443. [Google Scholar] [CrossRef]
- Iles, J.; Zmuidinaite, R.; Sadee, C.; Gardiner, A.; Lacey, J.; Harding, S.; Ule, J.; Roblett, D.; Heeney, J.; Baxendale, H.; et al. SARS-CoV-2 Spike Protein Binding of Glycated Serum Albumin—Its Potential Role in the Pathogenesis of the COVID-19 Clinical Syndromes and Bias towards Individuals with Pre-Diabetes/Type 2 Diabetes and Metabolic Diseases. Int. J. Mol. Sci. 2022, 23, 4126. [Google Scholar] [CrossRef] [PubMed]
- Iles, J.K.; Zmuidinaite, R.; Sadee, C.; Gardiner, A.; Lacey, J.; Harding, S.; Wallis, G.; Patel, R.; Roblett, D.; Heeney, J.; et al. Determination of IgG1 and IgG3 SARS-CoV-2 Spike Protein and Nucleocapsid Binding—Who Is Binding Who and Why? Int. J. Mol. Sci. 2022, 23, 6050. [Google Scholar] [CrossRef]
- Yates, J.L.; Ehrbar, D.J.; Hunt, D.T.; Girardin, R.C.; Dupuis, A.P., II; Payne, A.F.; Sowizral, M.; Varney, S.; Kulas, K.E.; Demarest, V.L.; et al. Serological Analysis Reveals an Imbalanced IgG Subclass Composition Associated with COVID-19 Disease Severity. medRxiv 2020. [Google Scholar] [CrossRef]
- Castillo-Olivares, J.; Wells, D.A.; Ferrari, M.; Chan, A.; Smith, P.; Nadesalingam, A.; Paloniemi, M.; Carnell, G.; Ohlendorf, L.; Cantoni, D.; et al. Towards Internationally standardised humoral Immune Correlates of Protection from SARS-CoV-2 infection and COVID-19 disease. medRxiv 2021. [Google Scholar] [CrossRef]
- Kussmann, M.; Roepstorff, P. Sample preparation techniques for peptides and proteins analyzed by MALDI-MS. Methods Mol. Biol. 2000, 146, 405–424. [Google Scholar] [PubMed]
- Strohalm, M.; Kavan, D.; Novak, P.; Volny, M.; Havlicek, V. mMass 3: A cross-platform software environment for precise analysis of mass spectrometric data. Anal. Chem. 2010, 82, 4648–4651. [Google Scholar] [CrossRef] [PubMed]
- The Lancet Diabetes Endocrinology. Editorial, COVID-19 and diabetes: A co-conspiracy? Lancet Diabetes Endocrinol. 2020, 8, 801. [Google Scholar] [CrossRef]
- Lauc, G.; Rudan, I.; Campbell, H.; Rudd, P.M. Complex genetic regulation of protein glycosylation. Mol Biosyst. 2010, 6, 329–335. [Google Scholar] [CrossRef] [Green Version]
- Ohtsubo, K.; Marth, J.D. Glycosylation in cellular mechanisms of health and disease. Cell 2006, 126, 855–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruhaak, L.R.; Zauner, G.; Huhn, C.; Bruggink, C.; Deelder, A.M.; Wuhrer, M. Glycan labeling strategies and their use in identification and quantification. Anal. Bioanal Chem 2010, 397, 3457–3481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guttman, A.; Chen, F.-T.A.; Evangelista, R.A.; Cooke, N. High-resolution capillary gel electrophoresis of reducing oligosaccharides labeled with 1-aminopyrene-3, 6, 8-trisulfonate. Anal. Biochem. 1996, 233, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.-T.A.; Dobashi, T.S.; Evangelista, R.A. Quantitative analysis of sugar constituents of glycoproteins by capillary electrophoresis. Glycobiology 1998, 8, 1045–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.; Zhang, L.; Thomas, S.; Hu, Y.; Li, S.; Cipollo, J.; Zhang, H. Modification of sialic acids on solid-phase: Accurate characterization of protein sialylation. Anal. Chem. 2017, 89, 6330–6335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.; Clark, D.; Liu, Y.; Li, S.; Zhang, H. High-throughput analysis of N-glycans using AutoTip via glycoprotein immobilization. Sci Rep. 2017, 7, 10216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, H.; Li, Y.; Sun, X.L. Recent advances in sialic acid-focused glycomics. J. Proteom. 2012, 75, 3098–3112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lattova, E.; Perreault, H.; Krokhin, O. Matrix-assisted laser desorption/ionization tandem mass spectrometry and post-source decay fragmentation study of phenylhydrazones of N-linked oligosaccharides from ovalbumin. J. Am. Soc. Mass Spectrom. 2004, 15, 725–735. [Google Scholar] [CrossRef] [Green Version]
- Remily-Wood, E.; Dirscherl, H.; Koomen, J.M. Acid Hydrolysis of Proteins in Matrix Assisted Laser Desorption Ionization Matrices. J. Am. Soc. Mass Spectrom. 2009, 20, 2106–2115. [Google Scholar] [CrossRef]
- Wide, L.; Eriksson, K.; Sluss, P.M.; Hall, J.E. Serum half-life of pituitary gonadotropins is decreased by sulfonation and increased by sialylation in women. J. Clin. Endocrinol. Metab. 2009, 94, 958–964. [Google Scholar] [CrossRef] [Green Version]
- Iles, R.K.; Chard, T. Molecular insights into the structure and function of human chorionic gonadotrophin. J. Mol. Endocrinol. 1993, 10, 217–234. [Google Scholar] [CrossRef] [PubMed]
- Furuhashi, M.; Shikone, T.; Fares, F.A.; Sugahara, T.; Hsueh, A.J.; Boime, I. Fusing the carboxy-terminal peptide of the chorionic gonadotropin (CG) beta-subunit to the common alpha-subunit: Retention of O-linked glycosylation and enhanced in vivo bioactivity of chimeric human CG. Mol. Endocrinol. 1995, 9, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Iles, R.K.; Cole, L.A.; Butler, S.A. Direct analysis of hCGβcf glycosylation in normal and aberrant pregnancy by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Int. J. Mol. Sci. 2014, 15, 10067–10082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plomp, R.; Dekkers, G.; Rombouts, Y.; Visser, R.; Koeleman, C.A.; Kammeijer, G.S.; Jansen, B.C.; Rispens, T.; Hensbergen, P.J.; Vidarsson, G.; et al. Hinge-Region O-Glycosylation of Human Immunoglobulin G3 (IgG3). Mol. Cell Proteom. 2015, 14, 1373–1384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medzihradszky, K.F.; Kaasik, K.; Chalkley, R.J. Tissue-Specific Glycosylation at the Glycopeptide Level. Mol. Cell Proteom. 2015, 14, 2103–2110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef]
- Stencel-Baerenwald, J.E.; Reiss, K.; Reiter, D.M.; Stehle, T.; Dermody, T.S. The sweet spot: Defining virus-sialic acid interactions. Nat. Rev. Microbiol. 2014, 12, 739–749. [Google Scholar] [CrossRef] [Green Version]
- Ayora-Talavera, G. Sialic acid receptors: Focus on their role in influenza infection. J. Recept. Ligand Channel Res. 2018, 10, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Hulswit, R.J.G.; Lang, Y.; Bakkers, M.J.G.; Li, W.; Li, Z.; Schouten, A.; Ophorst, B.; van Kuppeveld, F.J.M.; Boons, G.-J.; Bosch, B.-J.; et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl. Acad. Sci. USA 2019, 116, 2681–2690. [Google Scholar] [CrossRef]
- Kuchipudi, S.; Nelli, R.; Gontu, A.; Satyakumar, R.; Nair, M.S.; Subbiah, M. Sialic Acid Receptors: The Key to Solving the Enigma of Zoonotic Virus Spillover. Viruses 2021, 13, 262. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Petitjean, S.J.L.; Koehler, M.; Zhang, Q.; Dumitru, A.C.; Chen, W.; Derclaye, S.; Vincent, S.P.; Soumillion, P.; Alsteens, D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun 2020, 11, 4541. [Google Scholar] [CrossRef] [PubMed]
- Unione, L.; Moure, M.J.; Lenza, M.P.; Oyenarte, I.; Ereño-Orbea, J.; Ardá, A.; Jiménez-Barbero, J. The SARS‐CoV‐2 Spike Glycoprotein Directly Binds Exogeneous Sialic Acids: A NMR View. Angew. Chem. Int. Ed. 2022, 61, e202201432. [Google Scholar]
- Nguyen, L.; McCord, K.A.; Bui, D.T.; Bouwman, K.M.; Kitova, E.N.; Elaish, M.; Kumawat, D.; Daskhan, G.C.; Tomris, I.; Han, L.; et al. Sialic acid-containing glycolipids mediate binding and viral entry of SARS-CoV-2. Nat. Chem Biol 2022, 18, 81–90. [Google Scholar] [CrossRef]
- Seyran, M.; Takayama, K.; Uversky, V.N.; Lundstrom, K.; Palù, G.; Sherchan, S.P.; Attrish, D.; Rezaei, N.; Aljabali, A.A.A.; Ghosh, S.; et al. The structural basis of accelerated host cell entry by SARS-CoV-2. FEBS. J. 2021, 288, 5010–5020. [Google Scholar] [CrossRef]
- Baker, A.N.; Richards, S.-J.; Guy, C.S.; Congdon, T.R.; Hasan, M.; Zwetsloot, A.J.; Gallo, A.; Lewandowski, J.R.; Stansfeld, P.J.; Straube, A.; et al. The SARS-COV-2 Spike Protein Binds Sialic Acids and Enables Rapid Detection in a Lateral Flow Point of Care Diagnostic Device. ACS Cent. Sci. 2020, 6, 2046–2052. [Google Scholar] [CrossRef]
- Wu, J.T.; Tu, M.C.; Zhung, P. Advanced glycation end product (AGE): Characterization of the products from the reaction between D-glucose and serum albumin. J. Clin. Lab Anal. 1996, 10, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Cutler, D.M.; Summers, L.H. The COVID-19 Pandemic and the $16 Trillion Virus. JAMA 2020, 324, 1495–1496. [Google Scholar] [CrossRef]
- Available online: https://www.instituteforgovernment.org.uk/explainers/cost-coronavirus (accessed on 12 November 2021).
- Iles, R.K.; Zmuidinaite, R.; Iles, J.K.; Carnell, G.; Sampson, A.; Heeney, J.L. Development of a Clinical MALDI-ToF Mass Spectrometry Assay for SARS-CoV-2: Rational Design and Multi-Disciplinary Team Work. Diagnostics 2020, 10, 746. [Google Scholar] [CrossRef]
- Griffin, J.H.; Downard, K.M. Downard, Mass spectrometry analytical responses to the SARS-CoV-2 coronavirus in review. TrAC Trends Anal. Chem. 2021, 142, 116328. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iles, R.K.; Iles, J.K.; Lacey, J.; Gardiner, A.; Zmuidinaite, R. Direct Detection of Glycated Human Serum Albumin and Hyperglycosylated IgG3 in Serum, by MALDI-ToF Mass Spectrometry, as a Predictor of COVID-19 Severity. Diagnostics 2022, 12, 2521. https://doi.org/10.3390/diagnostics12102521
Iles RK, Iles JK, Lacey J, Gardiner A, Zmuidinaite R. Direct Detection of Glycated Human Serum Albumin and Hyperglycosylated IgG3 in Serum, by MALDI-ToF Mass Spectrometry, as a Predictor of COVID-19 Severity. Diagnostics. 2022; 12(10):2521. https://doi.org/10.3390/diagnostics12102521
Chicago/Turabian StyleIles, Ray K., Jason K. Iles, Jonathan Lacey, Anna Gardiner, and Raminta Zmuidinaite. 2022. "Direct Detection of Glycated Human Serum Albumin and Hyperglycosylated IgG3 in Serum, by MALDI-ToF Mass Spectrometry, as a Predictor of COVID-19 Severity" Diagnostics 12, no. 10: 2521. https://doi.org/10.3390/diagnostics12102521