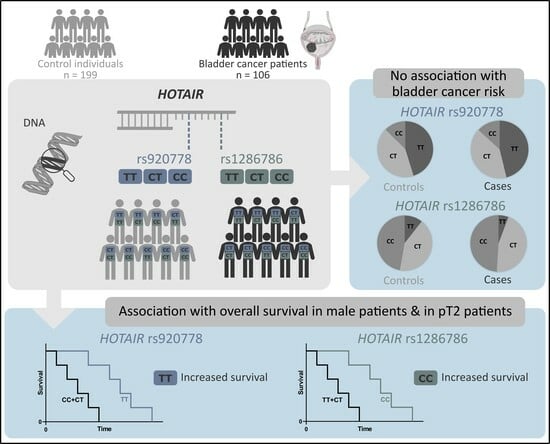
Relevance of HOTAIR rs920778 and rs12826786 Genetic Variants in Bladder Cancer Risk and Survival
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Single Nucleotide Polymorphism Genotyping
2.3. Statistical Analyses
3. Results
3.1. Population Characteristics
3.2. Genotype and Allelic Distributions of HOTAIR SNPs
3.3. HOTAIR rs920778 and rs12826786 SNPs and Bladder Cancer Risk
3.4. Effects of HOTAIR rs920778 and rs12826786 on Prognosis of Bladder Cancer Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA-Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Dyrskjøt, L.; Hansel, D.E.; Efstathiou, J.A.; Knowles, M.A.; Galsky, M.D.; Teoh, J.; Theodorescu, D. Bladder cancer. Nat. Rev. Dis. Primers 2023, 9, 58. [Google Scholar] [CrossRef] [PubMed]
- Babjuk, M.; Burger, M.; Capoun, O.; Cohen, D.; Compérat, E.M.; Escrig, J.L.D.; Gontero, P.; Liedberg, F.; Masson-Lecomte, A.; Mostafid, A.H. European Association of Urology guidelines on non–muscle-invasive bladder cancer (Ta, T1, and carcinoma in situ). Eur. Urol. 2022, 81, 75–94. [Google Scholar] [CrossRef]
- Witjes, J.A.; Bruins, H.M.; Cathomas, R.; Compérat, E.M.; Cowan, N.C.; Gakis, G.; Hernández, V.; Espinós, E.L.; Lorch, A.; Neuzillet, Y. European Association of Urology guidelines on muscle-invasive and metastatic bladder cancer: Summary of the 2020 guidelines. Eur. Urol. 2023, 79, 82–104. [Google Scholar] [CrossRef]
- Kaufman, D.S.; Shipley, W.U.; Feldman, A.S. Bladder cancer. Lancet 2009, 374, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Antoni, S.; Ferlay, J.; Soerjomataram, I.; Znaor, A.; Jemal, A.; Bray, F. Bladder cancer incidence and mortality: A global overview and recent trends. Eur. Urol. 2017, 71, 96–108. [Google Scholar] [CrossRef]
- Letašiová, S.; Medveďová, A.; Šovčíková, A.; Dušinská, M.; Volkovová, K.; Mosoiu, C.; Bartonová, A. Bladder cancer, a review of the environmental risk factors. Environ. Health 2012, 11, S11. [Google Scholar] [CrossRef]
- Cumberbatch, M.G.K.; Jubber, I.; Black, P.C.; Esperto, F.; Figueroa, J.D.; Kamat, A.M.; Kiemeney, L.; Lotan, Y.; Pang, K.; Silverman, D.T. Epidemiology of bladder cancer: A systematic review and contemporary update of risk factors in 2018. Eur. Urol. 2018, 74, 784–795. [Google Scholar] [CrossRef]
- Batista, R.; Vinagre, N.; Meireles, S.; Vinagre, J.; Prazeres, H.; Leão, R.; Máximo, V.; Soares, P. Biomarkers for bladder cancer diagnosis and surveillance: A comprehensive review. Diagnostics 2020, 10, 39. [Google Scholar] [CrossRef] [PubMed]
- Maas, M.; Todenhöfer, T.; Black, P.C. Urine biomarkers in bladder cancer—Current status and future perspectives. Nat. Rev. Urol. 2023, 20, 597–614. [Google Scholar] [CrossRef]
- Monreal-Trigo, J.; Alcañiz, M.; Martínez-Bisbal, M.C.; Loras, A.; Pascual, L.; Ruiz-Cerdá, J.L.; Ferrer, A.; Martínez-Máñez, R. New bladder cancer non-invasive surveillance method based on voltammetric electronic tongue measurement of urine. iScience 2022, 25, 104829. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.-C.; Hung, T.; Argani, P.; Rinn, J.L. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Xavier-Magalhães, A.; Gonçalves, C.; Fogli, A.; Lourenço, T.; Pojo, M.; Pereira, B.; Rocha, M.; Lopes, M.; Crespo, I.; Rebelo, O.; et al. The long non-coding RNA HOTAIR is transcriptionally activated by HOXA9 and is an independent prognostic marker in patients with malignant glioma. Oncotarget 2018, 9, 15740–15756. [Google Scholar] [CrossRef]
- Hajjari, M.; Salavaty, A. HOTAIR: An oncogenic long non-coding RNA in different cancers. Cancer Biol. Med. 2015, 12, 1–9. [Google Scholar] [CrossRef]
- Yu, X.; Li, Z. Long non-coding RNA HOTAIR: A novel oncogene. Mol. Med. Rep. 2015, 12, 5611–5618. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, L.; Wang, Y.; Li, H.; Ren, X.; Wei, F.; Yu, W.; Wang, X.; Zhang, L.; Yu, J. Long noncoding RNA HOTAIR involvement in cancer. Tumor Biol. 2014, 35, 9531–9538. [Google Scholar] [CrossRef]
- Rajagopal, T.; Talluri, S.; Akshaya, R.L.; Dunna, N.R. HOTAIR LncRNA: A novel oncogenic propellant in human cancer. Clin. Chim. Acta 2020, 503, 1–18. [Google Scholar] [CrossRef]
- Qu, X.; Alsager, S.; Zhuo, Y.; Shan, B. HOX transcript antisense RNA (HOTAIR) in cancer. Cancer Lett. 2019, 454, 90–97. [Google Scholar] [CrossRef]
- Raju, G.S.R.; Pavitra, E.; Bandaru, S.S.; Varaprasad, G.L.; Nagaraju, G.P.; Malla, R.R.; Huh, Y.S.; Han, Y.-K. HOTAIR: A potential metastatic, drug-resistant and prognostic regulator of breast cancer. Mol. Cancer 2023, 22, 65. [Google Scholar] [CrossRef]
- Zhu, C.; Wang, X.; Wang, Y.; Wang, K. Functions and underlying mechanisms of lncRNA HOTAIR in cancer chemotherapy resistance. Cell Death Discov. 2022, 8, 383. [Google Scholar] [CrossRef] [PubMed]
- Kogo, R.; Shimamura, T.; Mimori, K.; Kawahara, K.; Imoto, S.; Sudo, T.; Tanaka, F.; Shibata, K.; Suzuki, A.; Komune, S. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011, 71, 6320–6326. [Google Scholar] [CrossRef]
- Berrondo, C.; Flax, J.; Kucherov, V.; Siebert, A.; Osinski, T.; Rosenberg, A.; Fucile, C.; Richheimer, S.; Beckham, C.J. Expression of the long non-coding RNA HOTAIR correlates with disease progression in bladder cancer and is contained in bladder cancer patient urinary exosomes. PLoS ONE 2016, 11, e0147236. [Google Scholar] [CrossRef]
- Yan, T.-H.; Lu, S.-W.; Huang, Y.-Q.; Que, G.-B.; Chen, J.-H.; Chen, Y.-P.; Zhang, H.-B.; Liang, X.-L.; Jiang, J.-H. Upregulation of the long noncoding RNA HOTAIR predicts recurrence in stage Ta/T1 bladder cancer. Tumor Biol. 2014, 35, 10249–10257. [Google Scholar] [CrossRef]
- Tung, M.-C.; Wen, Y.-C.; Wang, S.-S.; Lin, Y.-W.; Chow, J.-M.; Yang, S.-F.; Chien, M.-H. Impact of long non-coding RNA HOTAIR genetic variants on the susceptibility and clinicopathologic characteristics of patients with urothelial cell carcinoma. J. Clin. Med. 2019, 8, 282. [Google Scholar] [CrossRef]
- Quan, J.; Pan, X.; Zhao, L.; Li, Z.; Dai, K.; Yan, F.; Liu, S.; Ma, H.; Lai, Y. LncRNA as a diagnostic and prognostic biomarker in bladder cancer: A systematic review and meta-analysis. OncoTargets Ther. 2018, 2018, 6415–6424. [Google Scholar] [CrossRef]
- Mao, X.; Chen, S.; Li, G. Identification of a ten-long noncoding RNA signature for predicting the survival and immune status of patients with bladder urothelial carcinoma based on the GEO database: A superior machine learning model. Aging 2021, 13, 6957. [Google Scholar] [CrossRef]
- Shang, C.; Guo, Y.; Zhang, H.; Xue, Y.-x. Long noncoding RNA HOTAIR is a prognostic biomarker and inhibits chemosensitivity to doxorubicin in bladder transitional cell carcinoma. Cancer Chemother. Pharmacol. 2016, 77, 507–513. [Google Scholar] [CrossRef]
- Martínez-Fernández, M.; Feber, A.; Dueñas, M.; Segovia, C.; Rubio, C.; Fernandez, M.; Villacampa, F.; Duarte, J.; López-Calderón, F.F.; Gómez-Rodriguez, M.J. Analysis of the Polycomb-related lncRNAs HOTAIR and ANRIL in bladder cancer. Clin. Epigenet. 2015, 7, 109. [Google Scholar] [CrossRef]
- Sun, X.; Du, P.; Yuan, W.; Du, Z.; Yu, M.; Yu, X.; Hu, T. Long non-coding RNA HOTAIR regulates cyclin J via inhibition of microRNA-205 expression in bladder cancer. Cell Death Dis. 2015, 6, e1907. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, W.; Zhang, Q.; Gu, D.; Zhang, K.; Ge, Y.; Chu, H.; Du, M.; Xu, B.; Wang, M. Tagging SNPs in the HOTAIR gene are associated with bladder cancer risk in a Chinese population. Gene 2018, 664, 22–26. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, L.; Fu, G.; Sun, F.; Shi, J.; Wei, J.; Lu, C.; Zhou, C.; Yuan, Q.; Yang, M. The identification of an ESCC susceptibility SNP rs920778 that regulates the expression of lncRNA HOTAIR via a novel intronic enhancer. Carcinogenesis 2014, 35, 2062–2067. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Dong, Z.; Bai, Y.; Guo, Y.; Shen, S.; Kuang, G.; Xu, J. Associations between polymorphisms of HOTAIR and risk of gastric cardia adenocarcinoma in a population of north China. Tumor Biol. 2015, 36, 2845–2854. [Google Scholar] [CrossRef]
- Pan, W.; Liu, L.; Wei, J.; Ge, Y.; Zhang, J.; Chen, H.; Zhou, L.; Yuan, Q.; Zhou, C.; Yang, M. A functional lncRNA HOTAIR genetic variant contributes to gastric cancer susceptibility. Mol. Carcinogen. 2016, 55, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Xavier-Magalhães, A.; Oliveira, A.I.; de Castro, J.V.; Pojo, M.; Gonçalves, C.S.; Lourenço, T.; Viana-Pereira, M.; Costa, S.; Linhares, P.; Vaz, R. Effects of the functional HOTAIR rs920778 and rs12826786 genetic variants in glioma susceptibility and patient prognosis. J. Neuro-Oncol. 2017, 132, 27–34. [Google Scholar] [CrossRef]
- Guo, L.; Lu, X.; Zheng, L.; Liu, X.; Hu, M. Association of long non-coding RNA HOTAIR polymorphisms with cervical cancer risk in a Chinese population. PLoS ONE 2016, 11, e0160039. [Google Scholar] [CrossRef] [PubMed]
- Min, L.; Mu, X.; Tong, A.; Qian, Y.; Ling, C.; Yi, T.; Zhao, X. The association between HOTAIR polymorphisms and cancer susceptibility: An updated systemic review and meta-analysis. OncoTargets Ther. 2018, 2018, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhao, Y.; Li, Y.; Lin, F.; Zhang, J. Association between HOTAIR genetic polymorphisms and cancer susceptibility: A meta-analysis involving 122,832 subjects. Genomics 2020, 112, 3036–3055. [Google Scholar] [CrossRef]
- WHO Classification of Tumours Editorial Board. Urinary and Male Genital Tumours—WHO Classification of Tumours, 5th ed.; IARC Press: Lyon, France, 2022; Volume 8. [Google Scholar]
- Oliveira, A.I.; Xavier-Magalhaes, A.; Moreira-Barbosa, C.; Magalhaes, H.; Henrique, R.; Jeronimo, C.; Costa, B.M. Influence of HOTAIR rs920778 and rs12826786 genetic variants on prostate cancer risk and progression-free survival. Biomark. Med. 2018, 12, 257–264. [Google Scholar] [CrossRef]
- Shi, Y.Y.; He, L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005, 15, 95–98. [Google Scholar] [CrossRef]
- Shariat, S.F.; Sfakianos, J.P.; Droller, M.J.; Karakiewicz, P.I.; Meryn, S.; Bochner, B.H. The effect of age and gender on bladder cancer: A critical review of the literature. BJU Int. 2010, 105, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Clements, J.A.; Batra, J. Single nucleotide polymorphisms in clinics: Fantasy or reality for cancer? Crit. Rev. Cl. Lab. Sci. 2016, 53, 29–39. [Google Scholar] [CrossRef]
- Estevão-Pereira, H.; Lobo, J.; Salta, S.; Amorim, M.; Lopes, P.; Cantante, M.; Reis, B.; Antunes, L.; Castro, F.; Palma de Sousa, S. Overexpression of circulating MiR-30b-5p identifies advanced breast cancer. J. Transl. Med. 2019, 17, 435. [Google Scholar] [CrossRef]
- Mugoni, V.; Ciani, Y.; Nardella, C.; Demichelis, F. Circulating RNAs in prostate cancer patients. Cancer Lett. 2022, 524, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Zhou, X.-y.; Du, X. Circulating long non-coding RNAs in cancer: Current status and future perspectives. Mol. Cancer 2016, 15, 39. [Google Scholar] [CrossRef] [PubMed]
- Franekova, M.; Halasova, E.; Bukovska, E.; Luptak, J.; Dobrota, D. Gene polymorphisms in bladder cancer. Urol. Oncol. 2008, 26, 1–8. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Dashti, S.; Farsi, M.; Taheri, M. HOX transcript antisense RNA: An oncogenic lncRNA in diverse malignancies. Exp. Mol. Pathol. 2021, 118, 104578. [Google Scholar] [CrossRef]
- Sørensen, K.P.; Thomassen, M.; Tan, Q.; Bak, M.; Cold, S.; Burton, M.; Larsen, M.J.; Kruse, T.A. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res. Treat. 2013, 142, 529–536. [Google Scholar] [CrossRef]
- Chen, F.J.; Sun, M.; Li, S.Q.; Wu, Q.Q.; Ji, L.; Liu, Z.L.; Zhou, G.Z.; Cao, G.; Jin, L.; Xie, H.W. Upregulation of the long non-coding rna hotair promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol. Carcinogen. 2013, 52, 908–915. [Google Scholar] [CrossRef]
- Ge, X.S.; Ma, H.J.; Zheng, X.H.; Ruan, H.L.; Liao, X.Y.; Xue, W.Q.; Chen, Y.B.; Zhang, Y.; Jia, W.H. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates W nt pathway. Cancer Sci. 2013, 104, 1675–1682. [Google Scholar] [CrossRef]
- Qi, Q.; Wang, J.; Huang, B.; Chen, A.; Li, G.; Li, X.; Wang, J. Association of HOTAIR polymorphisms rs4759314 and rs920778 with cancer susceptibility on the basis of ethnicity and cancer type. Oncotarget 2016, 7, 38775. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, G.; Guo, X.; Yao, H.; Wang, G.; Li, C. Non-coding RNA in bladder cancer. Cancer Lett. 2020, 485, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Aveta, A.; Cilio, S.; Contieri, R.; Spena, G.; Napolitano, L.; Manfredi, C.; Franco, A.; Crocerossa, F.; Cerrato, C.; Ferro, M. Urinary Micro-RNAs As Biomarkers of Urological Cancers: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 10846. [Google Scholar] [CrossRef] [PubMed]
- Hanke, M.; Hoefig, K.; Merz, H.; Feller, A.C.; Kausch, I.; Jocham, D.; Warnecke, J.M.; Sczakiel, G. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol. Oncol. 2010, 28, 655–661. [Google Scholar] [CrossRef]
- Shoemaker, R.; Kim, J. Urobiome: An outlook on the metagenome of urological diseases. Investig. Clin. Urol. 2021, 62, 611. [Google Scholar] [CrossRef]
- Nardelli, C.; Aveta, A.; Pandolfo, S.D.; Tripodi, L.; Russo, F.; Imbimbo, C.; Castaldo, G.; Pastore, L. Microbiome Profiling in Bladder Cancer Patients Using the First-morning Urine Sample. Eur. Urol. Open Sci. 2024, 59, 18–26. [Google Scholar] [CrossRef]

| Bladder Cancer Cases | Controls | |
|---|---|---|
| Number | 106 | 199 |
| Age, mean [range] | 67.35 [37–91] | 46.42 [27–85] |
| Sex, n (%) | ||
| Male | 83 (78.3%) | 130 (65.3%) |
| Female | 23 (21.7%) | 69 (34.7%) |
| Grade, n (%) | – | |
| PLG | 49 (46.2%) | |
| PHG | 30 (28.3%) | |
| IHG | 27 (25.5.%) | |
| Pathological Stage (pT), n (%) | – | |
| pTis | 4 (3.8%) | |
| pTa | 47 (44.3%) | |
| pT1 | 22 (20.8%) | |
| pT2 | 20 (18.9%) | |
| pT3 | 9 (8.5%) | |
| pT4 | 4 (3.8%) |
| Polymorphism | Controls | Cases | OR [95% CI] a | p-Value |
|---|---|---|---|---|
| HOTAIR rs920778 | ||||
| Genotype | ||||
| TT | 90 (45.2%) | 49 (46.2%) | - | 0.960 |
| CT | 84 (42.2%) | 43 (40.6%) | 0.940 [0.567–1.560] | 0.811 |
| CC | 25 (12.6%) | 14 (13.2%) | 1.029 [0.490–2.158] | 0.941 |
| CC+CT | 109 (54.8%) | 57 (53.8%) | 0.960 [0.599–1.541] | 0.867 |
| Alleles | ||||
| T | 264 (66.3%) | 141 (66.5%) | - | - |
| C | 134 (33.7%) | 71 (33.5%) | 0.992 [0.697–1.412] | 0.965 |
| HOTAIR rs12826786 | ||||
| Genotype | ||||
| CC | 94 (47.2%) | 52 (49.1%) | - | 0.367 |
| CT | 84 (42.2%) | 48 (45.3%) | 1.033 [0.633–1.687] | 0.897 |
| TT | 21 (10.6%) | 6 (5.7%) | 0.516 [0.196–1.360] | 0.181 |
| TT+CT | 105 (52.8%) | 54 (50.9%) | 0.930 [0.580–1.490] | 0.762 |
| Alleles | ||||
| C | 272 (68.3%) | 152 (71.7%) | - | - |
| T | 126 (31.7%) | 60 (28.3%) | 0.852 [0.591–1.229] | 0.391 |
| Polymorphism | Controls | Cases | OR [95% CI] a | p-Value |
|---|---|---|---|---|
| HOTAIR rs920778 | ||||
| Genotype | ||||
| TT | 90 (45.2%) | 49 (46.2%) | - | 0.690 |
| CT | 84 (42.2%) | 43 (40.6%) | 1.311 [0.664–2.588] | 0.435 |
| CC | 25 (12.6%) | 14 (13.2%) | 0.961 [0.373–2.477] | 0.935 |
| CC + CT | 109 (54.8%) | 57 (53.8%) | 1.207 [0.644–2.262] | 0.558 |
| Alleles | ||||
| T | 264 (66.3%) | 141 (66.5%) | - | - |
| C | 134 (33.7%) | 71 (33.5%) | 1.059 [0.667–1.682] | 0.807 |
| Age b | 1.127 [1.097–1.158] | <0.0001 | ||
| Sex | ||||
| Male | 130 (65.3%) | 83 (78.3%) | - | - |
| Female | 69 (34.7%) | 23 (21.7%) | 0.250 [0.118–0.529] | <0.001 |
| HOTAIR rs12826786 | ||||
| Genotype | ||||
| CC | 94 (47.2%) | 52 (49.1%) | - | 0.196 |
| CT | 84 (42.2%) | 48 (45.3%) | 1.359 [0.704–2.624] | 0.360 |
| TT | 21 (10.6%) | 6 (5.7%) | 0.439 [0.126–1.527] | 0.196 |
| TT + CT | 105 (52.8%) | 54 (50.9%) | 1.136 [0.608–2.124] | 0.689 |
| Alleles | ||||
| C | 272 (68.3%) | 152 (71.7%) | - | - |
| T | 126 (31.7%) | 60 (28.3%) | 0.913 [0.565–1.473] | 0.708 |
| Age b | 1.128 [1.098–1.159] | <0.0001 | ||
| Sex | ||||
| Male | 130 (65.3%) | 83 (78.3%) | - | - |
| Female | 69 (34.7%) | 23 (21.7%) | 0.235 [0.110–0.503] | <0.001 |
| Polymorphism | All Patients | HR [95% CI] a | p- Value | Male Patients | HR [95% CI] a | p- Value | pT2 Patients | HR [95% CI] a | p-Value |
|---|---|---|---|---|---|---|---|---|---|
| HOTAIRrs920778 | |||||||||
| Genotype | |||||||||
| TT | 49 | - | 0.322 | 38 | - | 0.086 | 8 | - | 0.111 |
| CT | 43 | 1.484 [0.783–2.812] | 0.226 | 34 | 1.950 [0.936–4.062] | 0.075 | 9 | 4.248 [1.026–17.589] | 0.046 |
| CC | 14 | 1.924 [0.716–5.173] | 0.195 | 11 | 2.841 [0.975–8.278] | 0.056 | 3 | 4.497 [0.812–24.890] | 0.085 |
| CC+CT | 57 | 1.550 [0.840–2.858] | 0.161 | 45 | 2.098 [1.047–4.204] | 0.037 | 12 | 4.313 [1.099–16.931] | 0.036 |
| Age at diagnosis b | 1.051 [1.014–1.090] | 0.007 | 1.041 [0.999–1.083] | 0.053 | 1.014 [0.962–1.069] | 0.600 | |||
| Sex | - | - | - | ||||||
| Male | 83 | - | - | - | - | - | 18 | - | - |
| Female | 23 | 0.838 [0.378–1.858] | 0.664 | - | - | - | 2 | 1.271 [0.132–12.218] | 0.835 |
| Pathological stage (pT) | - | - | - | ||||||
| pTis | 4 | - | <0.0001 | 4 | - | <0.0001 | - | - | - |
| pTa | 47 | 0.801 [0.102–6.285] | 0.833 | 35 | 0.619 [0.077–5.001] | 0.652 | - | - | - |
| pT1 | 22 | 1.067 [0.128–8.918] | 0.953 | 14 | 1.230 [0.144–10.541] | 0.850 | - | - | - |
| pT2 | 20 | 3.027 [0.391–23.414] | 0.289 | 18 | 3.075 [0.394–23.982] | 0.284 | - | - | - |
| pT3 | 9 | 2.765 [0.318–24.022] | 0.357 | 8 | 2.928 [0.320–26.790] | 0.341 | - | - | - |
| pT4 | 4 | 40.661 [4.095–403.734] | 0.002 | 4 | 39.680 [3.906–403.059] | 0.002 | - | - | - |
| HOTAIRrs12826786 | |||||||||
| Genotype | |||||||||
| CC | 52 | - | 0.198 | 41 | - | 0.038 | 7 | - | 0.043 |
| CT | 48 | 1.574 [0.837–2.960] | 0.159 | 37 | 2.332 [1.136–4.790] | 0.021 | 11 | 6.343 [1.303–30.883] | 0.022 |
| TT | 6 | 2.547 [0.776–8.365] | 0.123 | 5 | 3.384 [0.936–12.236] | 0.063 | 2 | 11.504 [1.366–96.901] | 0.025 |
| TT+CT | 54 | 1.644 [0.888–3.045] | 0.114 | 42 | 2.432 [1.207–4.898] | 0.013 | 13 | 6.788 [1.427–32.284] | 0.016 |
| Age at diagnosis b | 1.051 [1.013–1.091] | 0.008 | 1.036 [0.995–1.078] | 0.089 | 0.995 [0.938–1.055] | 0.868 | |||
| Sex | - | - | - | ||||||
| Male | 83 | - | - | - | - | - | 18 | - | - |
| Female | 23 | 0.815 [0.369–1.800] | 0.612 | - | - | - | 2 | 0.437 [0.049–3.887] | 0.458 |
| Pathological stage (pT) | - | - | - | ||||||
| pTis | 4 | - | <0.0001 | 4 | - | <0.0001 | - | - | - |
| pTa | 47 | 0.847 [0.108–6.625] | 0.874 | 35 | 0.639 [0.079–5.180] | 0.675 | - | - | - |
| pT1 | 22 | 1.112 [0.133–9.330] | 0.922 | 14 | 1.320 [0.154–11.270] | 0.800 | - | - | - |
| pT2 | 20 | 2.962 [0.385–22.799] | 0.297 | 18 | 3.245 [0.419–25.134] | 0.260 | - | - | - |
| pT3 | 9 | 2.553 [0.289–22.565] | 0.399 | 8 | 2.680 [0.285–25.204] | 0.389 | - | - | - |
| pT4 | 4 | 41.616 [4.177–414.679] | 0.001 | 4 | 38.968 [3.831–396.417] | 0.002 | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martins, E.P.; Vieira de Castro, J.; Fontes, R.; Monteiro-Reis, S.; Henrique, R.; Jerónimo, C.; Costa, B.M. Relevance of HOTAIR rs920778 and rs12826786 Genetic Variants in Bladder Cancer Risk and Survival. Cancers 2024, 16, 434. https://doi.org/10.3390/cancers16020434
Martins EP, Vieira de Castro J, Fontes R, Monteiro-Reis S, Henrique R, Jerónimo C, Costa BM. Relevance of HOTAIR rs920778 and rs12826786 Genetic Variants in Bladder Cancer Risk and Survival. Cancers. 2024; 16(2):434. https://doi.org/10.3390/cancers16020434
Chicago/Turabian StyleMartins, Eduarda P., Joana Vieira de Castro, Rita Fontes, Sara Monteiro-Reis, Rui Henrique, Carmen Jerónimo, and Bruno M. Costa. 2024. "Relevance of HOTAIR rs920778 and rs12826786 Genetic Variants in Bladder Cancer Risk and Survival" Cancers 16, no. 2: 434. https://doi.org/10.3390/cancers16020434