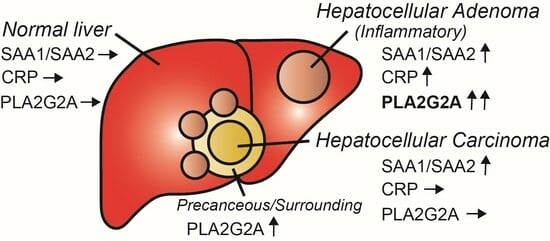
Phospholipase A2 Group IIA Is Associated with Inflammatory Hepatocellular Adenoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Characteristics of the Patient
2.2. Human Liver Tissue Dissociation and Single-Cell RNA Sequencing (scRNA-seq)
2.3. Immunohistochemistry (IHC)
2.4. RNA Sequencing (RNA-seq)
3. Results
3.1. scRNA-seq of HCA Sample
3.2. Comparison with Other HCC Samples by scRNA-seq Analysis
3.3. IHC of PLA2G2A in I-HCA Sample
3.4. Distribution of PLA2G2A Expression in Normal and HBV-HCC Samples
3.5. Bulk RNA-seq Analysis for Two Different I-HCA Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bioulac-Sage, P.; Taouji, S.; Possenti, L.; Balabaud, C. Hepatocellular adenoma subtypes: The impact of overweight and obesity. Liver Int. 2012, 32, 1217–1221. [Google Scholar] [CrossRef]
- Dokmak, S.; Paradis, V.; Vilgrain, V.; Sauvanet, A.; Farges, O.; Valla, D.; Bedossa, P.; Belghiti, J. A single-center surgical experience of 122 patients with single and multiple hepatocellular adenomas. Gastroenterology 2009, 137, 1698–1705. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Nakanuma, Y. Overview of hepatocellular adenoma in Japan. Int. J. Hepatol. 2012, 2012, 648131. [Google Scholar] [CrossRef] [PubMed]
- Nault, J.C.; Couchy, G.; Balabaud, C.; Morcrette, G.; Caruso, S.; Blanc, J.F.; Bacq, Y.; Calderaro, J.; Paradis, V.; Ramos, J.; et al. Molecular Classification of Hepatocellular Adenoma Associates with Risk Factors, Bleeding, and Malignant Transformation. Gastroenterology 2017, 152, 880–894.e6. [Google Scholar] [CrossRef]
- Bioulac-Sage, P.; Gouw, A.S.H.; Balabaud, C.; Sempoux, C. Hepatocellular adenoma: What we know, what we do not know, and why it matters. Histopathology 2022, 80, 878–897. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, X. Hepatocellular adenoma: Where are we now? World J. Gastroenterol. 2022, 28, 1384–1393. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, S. Nx1-Seq (Well Based Single-Cell Analysis System). Adv. Exp. Med. Biol. 2019, 1129, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, S.; Tabuchi, Y.; Yurino, H.; Hirohashi, Y.; Deshimaru, S.; Asano, T.; Mariya, T.; Oshima, K.; Takamura, Y.; Ukita, Y.; et al. Comprehensive single-cell transcriptome analysis reveals heterogeneity in endometrioid adenocarcinoma tissues. Sci. Rep. 2017, 7, 14225. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Zhu, C.; Song, H.; Shen, B.; Wu, L.; Liu, F.; Liu, X. Promoting effect of hepatitis B virus on the expressoin of phospholipase A2 group IIA. Lipids Health Dis. 2017, 16, 5. [Google Scholar] [CrossRef]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Robinson, B.S.; Hii, C.S.; Ferrante, A. Activation of phospholipase A2 in human neutrophils by polyunsaturated fatty acids and its role in stimulation of superoxide production. Biochem. J. 1998, 336 Pt 3, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Taketomi, Y.; Girard, C.; Yamamoto, K.; Lambeau, G. Emerging roles of secreted phospholipase A2 enzymes: Lessons from transgenic and knockout mice. Biochimie 2010, 92, 561–582. [Google Scholar] [CrossRef] [PubMed]
- Mauchley, D.; Meng, X.; Johnson, T.; Fullerton, D.A.; Weyant, M.J. Modulation of growth in human esophageal adenocarcinoma cells by group IIa secretory phospholipase A(2). J. Thorac. Cardiovasc. Surg. 2010, 139, 591–599; discussion 599. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.K.; Rostomily, R.; Wong, S.T.C. Prognostic Gene Discovery in Glioblastoma Patients using Deep Learning. Cancers 2019, 11, 53. [Google Scholar] [CrossRef] [PubMed]
- He, H.L.; Lee, Y.E.; Shiue, Y.L.; Lee, S.W.; Lin, L.C.; Chen, T.J.; Wu, T.F.; Li, C.F. PLA2G2A overexpression is associated with poor therapeutic response and inferior outcome in rectal cancer patients receiving neoadjuvant concurrent chemoradiotherapy. Histopathology 2015, 66, 991–1002. [Google Scholar] [CrossRef]
- Zhang, M.; Xiang, R.; Glorieux, C.; Huang, P. PLA2G2A Phospholipase Promotes Fatty Acid Synthesis and Energy Metabolism in Pancreatic Cancer Cells with K-ras Mutation. Int. J. Mol. Sci. 2022, 23, 11721. [Google Scholar] [CrossRef]
- Leung, S.Y.; Chen, X.; Chu, K.M.; Yuen, S.T.; Mathy, J.; Ji, J.; Chan, A.S.; Li, R.; Law, S.; Troyanskaya, O.G.; et al. Phospholipase A2 group IIA expression in gastric adenocarcinoma is associated with prolonged survival and less frequent metastasis. Proc. Natl. Acad. Sci. USA 2002, 99, 16203–16208. [Google Scholar] [CrossRef]
- Ganesan, K.; Ivanova, T.; Wu, Y.; Rajasegaran, V.; Wu, J.; Lee, M.H.; Yu, K.; Rha, S.Y.; Chung, H.C.; Ylstra, B.; et al. Inhibition of gastric cancer invasion and metastasis by PLA2G2A, a novel beta-catenin/TCF target gene. Cancer Res. 2008, 68, 4277–4286. [Google Scholar] [CrossRef]
- Xing, X.F.; Li, H.; Zhong, X.Y.; Zhang, L.H.; Wang, X.H.; Liu, Y.Q.; Jia, S.Q.; Shi, T.; Niu, Z.J.; Peng, Y.; et al. Phospholipase A2 group IIA expression correlates with prolonged survival in gastric cancer. Histopathology 2011, 59, 198–206. [Google Scholar] [CrossRef]
- Shariati, M.; Aghaei, M.; Movahedian, A.; Somi, M.H.; Dolatkhah, H.; Aghazade, A.M. The effect of omega-fatty acids on the expression of phospholipase A2 group 2A in human gastric cancer patients. J. Res. Med. Sci. 2016, 21, 10. [Google Scholar] [CrossRef] [PubMed]
- Menschikowski, M.; Hagelgans, A.; Schuler, U.; Froeschke, S.; Rosner, A.; Siegert, G. Plasma levels of phospholipase A2-IIA in patients with different types of malignancies: Prognosis and association with inflammatory and coagulation biomarkers. Pathol. Oncol. Res. 2013, 19, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Ponten, F.; Jirstrom, K.; Uhlen, M. The Human Protein Atlas—A tool for pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Lanczky, A.; Gyorffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
- Rebouissou, S.; Franconi, A.; Calderaro, J.; Letouze, E.; Imbeaud, S.; Pilati, C.; Nault, J.C.; Couchy, G.; Laurent, A.; Balabaud, C.; et al. Genotype-phenotype correlation of CTNNB1 mutations reveals different ss-catenin activity associated with liver tumor progression. Hepatology 2016, 64, 2047–2061. [Google Scholar] [CrossRef]
- Cao, W.; Li, M.; Liu, J.; Zhang, S.; Noordam, L.; Verstegen, M.M.A.; Wang, L.; Ma, B.; Li, S.; Wang, W.; et al. LGR5 marks targetable tumor-initiating cells in mouse liver cancer. Nat. Commun. 2020, 11, 1961. [Google Scholar] [CrossRef]
- He, J.; Han, J.; Lin, K.; Wang, J.; Li, G.; Li, X.; Gao, Y. PTEN/AKT and Wnt/beta-catenin signaling pathways regulate the proliferation of Lgr5+ cells in liver cancer. Biochem. Biophys. Res. Commun. 2023, 683, 149117. [Google Scholar] [CrossRef]
- Han, J.; Lin, K.; Zhang, X.; Yan, L.; Chen, Y.; Chen, H.; Liu, J.; Liu, J.; Wu, Y. PTEN-mediated AKT/beta-catenin signaling enhances the proliferation and expansion of Lgr5+ hepatocytes. Int. J. Biol. Sci. 2021, 17, 861–868. [Google Scholar] [CrossRef]
- Suppli, M.P.; Rigbolt, K.T.G.; Veidal, S.S.; Heeboll, S.; Eriksen, P.L.; Demant, M.; Bagger, J.I.; Nielsen, J.C.; Oro, D.; Thrane, S.W.; et al. Hepatic transcriptome signatures in patients with varying degrees of nonalcoholic fatty liver disease compared with healthy normal-weight individuals. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 316, G462–G472. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iwabuchi, S.; Takahashi, K.; Kawaguchi, K.; Nagatsu, A.; Imafuku, T.; Shichino, S.; Matsushima, K.; Taketomi, A.; Honda, M.; Hashimoto, S. Phospholipase A2 Group IIA Is Associated with Inflammatory Hepatocellular Adenoma. Cancers 2024, 16, 159. https://doi.org/10.3390/cancers16010159
Iwabuchi S, Takahashi K, Kawaguchi K, Nagatsu A, Imafuku T, Shichino S, Matsushima K, Taketomi A, Honda M, Hashimoto S. Phospholipase A2 Group IIA Is Associated with Inflammatory Hepatocellular Adenoma. Cancers. 2024; 16(1):159. https://doi.org/10.3390/cancers16010159
Chicago/Turabian StyleIwabuchi, Sadahiro, Kenta Takahashi, Kazunori Kawaguchi, Akihisa Nagatsu, Tadashi Imafuku, Shigeyuki Shichino, Kouji Matsushima, Akinobu Taketomi, Masao Honda, and Shinichi Hashimoto. 2024. "Phospholipase A2 Group IIA Is Associated with Inflammatory Hepatocellular Adenoma" Cancers 16, no. 1: 159. https://doi.org/10.3390/cancers16010159