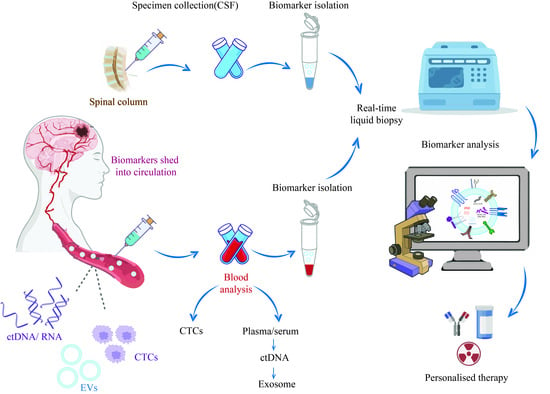
The Current Landscape of Glioblastoma Biomarkers in Body Fluids
Abstract
:Simple Summary
Abstract
1. Introduction
2. Circulating Tumor Cells as Glioblastoma Biomarkers
3. Cell-Free Nucleic Acids as Glioblastoma Biomarkers
3.1. Cell-Free DNA
3.2. Cell-Free RNA
4. Extracellular Vesicles as Glioblastoma Biomarkers
4.1. Exosomes
4.2. Microvesicles
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sasmita, A.O.; Wong, Y.P.; Ling, A.P.K. Biomarkers and therapeutic advances in glioblastoma multiforme. Asia-Pac. J. Clin. Oncol. 2018, 14, 40–51. [Google Scholar] [CrossRef] [Green Version]
- Mair, R.; Mouliere, F. Cell-free DNA technologies for the analysis of brain cancer. Br. J. Cancer 2022, 126, 371–378. [Google Scholar] [CrossRef] [PubMed]
- King, J.L.; Benhabbour, S.R. Glioblastoma Multiforme—A Look at the Past and a Glance at the Future. Pharmaceutics 2021, 13, 1053. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhou, W.; Tan, Y.; Tian, D.; Zhong, C. 5-Hydroxymethylcytosines in circulating cell-free DNA reveal a diagnostic biomarker for glioma. Heliyon 2022, 8, e11022. [Google Scholar] [CrossRef]
- King, J.L.; Maturavongsadit, P.; Hingtgen, S.D.; Benhabbour, S.R. Injectable pH Thermo-Responsive Hydrogel Scaffold for Tumoricidal Neural Stem Cell Therapy for Glioblastoma Multiforme. Pharmaceutics 2022, 14, 2243. [Google Scholar] [CrossRef]
- Müller Bark, J.; Kulasinghe, A.; Chua, B.; Day, B.W.; Punyadeera, C. Circulating biomarkers in patients with glioblastoma. Br. J. Cancer 2020, 122, 295–305. [Google Scholar] [CrossRef] [Green Version]
- Khristov, V.; Lin, A.; Freedman, Z.; Staub, J.; Shenoy, G.; Mrowczynski, O.; Rizk, E.; Zacharia, B.; Connor, J. Tumor-derived Biomarkers in Liquid Biopsy of Glioblastoma. World Neurosurg. 2022, 170, 182–194. [Google Scholar] [CrossRef]
- Stupp, R.; Brada, M.; Van Den Bent, M.; Tonn, J.-C.; Pentheroudakis, G. High-grade glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2014, 25, iii93–iii101. [Google Scholar] [PubMed]
- Best, M.G.; Sol, N.; Zijl, S.; Reijneveld, J.C.; Wesseling, P.; Wurdinger, T. Liquid biopsies in patients with diffuse glioma. Acta Neuropathol. 2015, 129, 849–865. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Bettegowda, C. Applications of DNA-based liquid biopsy for central nervous system neoplasms. J. Mol. Diagn. 2017, 19, 24–34. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Shan, S.; Xia, B.; Zhang, L.; Liang, X.J. Brain-Targeted Exosomes-Based Drug Delivery System to Overcome the Treatment Bottleneck of Brainstem Glioma. Adv. Funct. Mater. 2023, 5, 2302378. [Google Scholar]
- Xiao, F.; Lv, S.; Zong, Z.; Wu, L.; Tang, X.; Kuang, W.; Zhang, P.; Li, X.; Fu, J.; Xiao, M. Cerebrospinal fluid biomarkers for brain tumor detection: Clinical roles and current progress. Am. J. Transl. Res. 2020, 12, 1379. [Google Scholar] [PubMed]
- Westphal, M.; Lamszus, K. Circulating biomarkers for gliomas. Nat. Rev. Neurol. 2015, 11, 556–566. [Google Scholar] [CrossRef]
- Jelski, W.; Mroczko, B. Molecular and circulating biomarkers of brain tumors. Int. J. Mol. Sci. 2021, 22, 7039. [Google Scholar] [CrossRef]
- Caputo, V.; Ciardiello, F.; Della Corte, C.M.; Martini, G.; Troiani, T.; Napolitano, S. Diagnostic value of liquid biopsy in the era of precision medicine: 10 years of clinical evidence in cancer. Explor. Target. Anti-Tumor Ther. 2023, 4, 102. [Google Scholar]
- Adams, E.; Sepich-Poore, G.D.; Miller-Montgomery, S.; Knight, R. Using all our genomes: Blood-based liquid biopsies for the early detection of cancer. View 2022, 3, 20200118. [Google Scholar]
- Shen, C.-I.; Chiang, C.-L.; Shiao, T.-H.; Luo, Y.-H.; Chao, H.-S.; Huang, H.-C.; Chiu, C.-H. Real-world evidence of the intrinsic limitations of PCR-based EGFR mutation assay in non-small cell lung cancer. Sci. Rep. 2022, 12, 13566. [Google Scholar]
- Lamb, Y.N.; Dhillon, S. Epi proColon® 2.0 CE: A blood-based screening test for colorectal cancer. Mol. Diagn. Ther. 2017, 21, 225–232. [Google Scholar] [CrossRef]
- Yi, Z.; Qu, C.; Zeng, Y.; Liu, Z. Liquid biopsy: Early and accurate diagnosis of brain tumor. J. Cancer Res. Clin. Oncol. 2022, 148, 2347–2373. [Google Scholar]
- An, Y.; Fan, F.; Jiang, X.; Sun, K. Recent advances in liquid biopsy of brain cancers. Front. Genet. 2021, 12, 720270. [Google Scholar] [CrossRef]
- Gatto, L.; Franceschi, E.; Di Nunno, V.; Tosoni, A.; Lodi, R.; Brandes, A.A. Liquid biopsy in glioblastoma management: From current research to future perspectives. Oncology 2021, 26, 865–878. [Google Scholar]
- Saenz-Antoñanzas, A.; Auzmendi-Iriarte, J.; Carrasco-Garcia, E.; Moreno-Cugnon, L.; Ruiz, I.; Villanua, J.; Egaña, L.; Otaegui, D.; Samprón, N.; Matheu, A. Liquid biopsy in glioblastoma: Opportunities, applications and challenges. Cancers 2019, 11, 950. [Google Scholar] [CrossRef] [Green Version]
- Tamai, S.; Ichinose, T.; Nakada, M. Liquid biomarkers in glioma. Brain Tumor Pathol. 2023, 40, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yuan, F.; Qi, Y.; Liu, B.; Chen, Q. Circulating tumor cells for glioma. Front. Oncol. 2021, 11, 607150. [Google Scholar] [CrossRef] [PubMed]
- Lynch, D.; Powter, B.; Po, J.W.; Cooper, A.; Garrett, C.; Koh, E.-S.; Sheridan, M.; van Gelder, J.; Darwish, B.; Mckechnie, S. Isolation of circulating tumor cells from glioblastoma patients by direct immunomagnetic targeting. Appl. Sci. 2020, 10, 3338. [Google Scholar] [CrossRef]
- MacArthur, K.M.; Kao, G.D.; Chandrasekaran, S.; Alonso-Basanta, M.; Chapman, C.; Lustig, R.A.; Wileyto, E.P.; Hahn, S.M.; Dorsey, J.F. Detection of brain tumor cells in the peripheral blood by a telomerase promoter-based assay. Cancer Res. 2014, 74, 2152–2159. [Google Scholar] [CrossRef] [Green Version]
- Müller, C.; Holtschmidt, J.; Auer, M.; Heitzer, E.; Lamszus, K.; Schulte, A.; Matschke, J.; Langer-Freitag, S.; Gasch, C.; Stoupiec, M. Hematogenous dissemination of glioblastoma multiforme. Sci. Transl. Med. 2014, 6, ra101–ra247. [Google Scholar] [CrossRef]
- Sullivan, J.P.; Nahed, B.V.; Madden, M.W.; Oliveira, S.M.; Springer, S.; Bhere, D.; Chi, A.S.; Wakimoto, H.; Rothenberg, S.M.; Sequist, L.V. Brain tumor cells in circulation are enriched for mesenchymal gene expression. Cancer Discov. 2014, 4, 1299–1309. [Google Scholar] [CrossRef] [Green Version]
- Chistiakov, D.A.; Chekhonin, V.P. Circulating tumor cells and their advances to promote cancer metastasis and relapse, with focus on glioblastoma multiforme. Exp. Mol. Pathol. 2018, 105, 166–174. [Google Scholar]
- Touat, M.; Duran-Peña, A.; Alentorn, A.; Lacroix, L.; Massard, C.; Idbaih, A. Emerging circulating biomarkers in glioblastoma: Promises and challenges. Expert Rev. Mol. Diagn. 2015, 15, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Sabath, D.E.; Perrone, M.E.; Clein, A.; Tam, M.; Hardin, M.; Trimble, S.; Ramirez, A.; Duplessis, M.; Mojica, T.; Higano, C.S. Clinical Validation of a Circulating Tumor Cell Assay Using Density Centrifugation and Automated Immunofluorescence Microscopy. Am. J. Clin. Pathol. 2022, 158, 270–276. [Google Scholar] [CrossRef]
- Kumaria, A.; Teale, A.; Kulkarni, G.V.; Ingale, H.A.; Macarthur, D.C.; Robertson, I.J. Glioblastoma multiforme metastatic to lung in the absence of intracranial recurrence: Case report. Br. J. Neurosurg. 2022, 36, 290–292. [Google Scholar] [CrossRef]
- Strong, M.J.; Koduri, S.; Allison, J.A.; Pesavento, C.M.; Ogunsola, S.; Ogunsola, O.; Yee, T.J.; Khalsa, S.S.S.; Saadeh, Y.S.; Joseph, J.R. Bone metastasis from glioblastoma: A systematic review. J. Neurooncol. 2022, 158, 379–392. [Google Scholar] [CrossRef]
- Laguado, M.Z.; Baez, J.M.; Luna, A.; Mantilla, C.; Palencia, M.; Pimiento, J.M.B.; Luna-Meza, A.; Palencia Sr, M. Bone metastasis from glioblastoma multiforme: A case report. Cureus 2022, 14, e25464. [Google Scholar]
- Artzi, M.; Bressler, I.; Ben Bashat, D. Differentiation between glioblastoma, brain metastasis and subtypes using radiomics analysis. J. Magn. Reson. Imaging 2019, 50, 519–528. [Google Scholar] [CrossRef]
- Caramanti, R.L.; Aprígio, R.M.; Tognola, W.A.; Laurenti, M.R.; Rocha, C.E.; Góes, M.J. Transtentorial spread of glioblastoma multiforme to cerebellopontine angle–A rare case report. Surg. Neurol. Int. 2022, 13, 8813625. [Google Scholar] [CrossRef]
- Cirkel, G.A.; Gadellaa-van Hooijdonk, C.G.; Koudijs, M.J.; Willems, S.M.; Voest, E.E. Tumor heterogeneity and personalized cancer medicine: Are we being outnumbered? Future Oncol. 2014, 10, 417–428. [Google Scholar] [CrossRef]
- Fontanilles, M.; Sanson, M.; Touat, M. Liquid biopsy in neuro-oncology: Are we finally there? Ann. Oncol. 2021, 32, 1472–1474. [Google Scholar] [CrossRef]
- Palande, V.; Siegal, T.; Detroja, R.; Gorohovski, A.; Glass, R.; Flueh, C.; Kanner, A.A.; Laviv, Y.; Har-Nof, S.; Levy-Barda, A. Detection of gene mutations and gene–gene fusions in circulating cell-free DNA of glioblastoma patients: An avenue for clinically relevant diagnostic analysis. Mol. Oncol. 2022, 16, 2098–2114. [Google Scholar]
- Li, D.; Bonner, E.R.; Wierzbicki, K.; Panditharatna, E.; Huang, T.; Lulla, R.; Mueller, S.; Koschmann, C.; Nazarian, J.; Saratsis, A.M. Standardization of the liquid biopsy for pediatric diffuse midline glioma using ddPCR. Sci. Rep. 2021, 11, 5098. [Google Scholar] [CrossRef]
- Johnson, K.C.; Verhaak, R.G. Serum cell-free DNA epigenetic biomarkers aid glioma diagnostics and monitoring. Neuro-Oncology 2021, 23, 1423–1424. [Google Scholar] [CrossRef]
- Nabavizadeh, S.A.; Ware, J.B.; Guiry, S.; Nasrallah, M.P.; Mays, J.J.; Till, J.E.; Hussain, J.; Abdalla, A.; Yee, S.S.; Binder, Z.A. Imaging and histopathologic correlates of plasma cell-free DNA concentration and circulating tumor DNA in adult patients with newly diagnosed glioblastoma. Neuro-Oncol. Adv. 2020, 2, vdaa016. [Google Scholar] [CrossRef]
- Bagley, S.J.; Nabavizadeh, S.A.; Mays, J.J.; Till, J.E.; Ware, J.B.; Levy, S.; Sarchiapone, W.; Hussain, J.; Prior, T.; Guiry, S. Clinical Utility of Plasma Cell-Free DNA in Adult Patients with Newly Diagnosed Glioblastoma: A Pilot Prospective StudyPlasma cfDNA in Glioblastoma. Clin. Cancer Res. 2020, 26, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Fontanilles, M.; Marguet, F.; Beaussire, L.; Magne, N.; Pépin, L.-F.; Alexandru, C.; Tennevet, I.; Hanzen, C.; Langlois, O.; Jardin, F. Cell-free DNA and circulating TERT promoter mutation for disease monitoring in newly-diagnosed glioblastoma. Acta Neuropathol. Commun. 2020, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nørøxe, D.S.; Østrup, O.; Yde, C.W.; Ahlborn, L.B.; Nielsen, F.C.; Michaelsen, S.R.; Larsen, V.A.; Skjøth-Rasmussen, J.; Brennum, J.; Hamerlik, P. Cell-free DNA in newly diagnosed patients with glioblastoma–a clinical prospective feasibility study. Oncotarget 2019, 10, 4397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagley, S.J.; Till, J.; Abdalla, A.; Sangha, H.K.; Yee, S.S.; Freedman, J.; Black, T.A.; Hussain, J.; Binder, Z.A.; Brem, S. Association of plasma cell-free DNA with survival in patients with IDH wild-type glioblastoma. Neuro-Oncol. Adv. 2021, 3, vdab011. [Google Scholar] [CrossRef]
- Mouliere, F.; Smith, C.G.; Heider, K.; Su, J.; van der Pol, Y.; Thompson, M.; Morris, J.; Wan, J.C.; Chandrananda, D.; Hadfield, J. Fragmentation patterns and personalized sequencing of cell-free DNA in urine and plasma of glioma patients. EMBO Mol. Med. 2021, 13, e12881. [Google Scholar] [CrossRef]
- Fontanilles, M.; Marguet, F.; Bohers, É.; Viailly, P.-J.; Dubois, S.; Bertrand, P.; Camus, V.; Mareschal, S.; Ruminy, P.; Maingonnat, C. Non-invasive detection of somatic mutations using next-generation sequencing in primary central nervous system lymphoma. Oncotarget 2017, 8, 48157. [Google Scholar] [CrossRef] [Green Version]
- Liebs, S.; Eder, T.; Klauschen, F.; Schütte, M.; Yaspo, M.-L.; Keilholz, U.; Tinhofer, I.; Kidess-Sigal, E.; Braunholz, D. Applicability of liquid biopsies to represent the mutational profile of tumor tissue from different cancer entities. Oncogene 2021, 40, 5204–5212. [Google Scholar] [CrossRef]
- Ramkissoon, L.A.; Pegram, W.; Haberberger, J.; Danziger, N.; Lesser, G.; Strowd, R.; Dahiya, S.; Cummings, T.J.; Bi, W.L.; Abedalthagafi, M. Genomic profiling of circulating tumor DNA from cerebrospinal fluid to guide clinical decision making for patients with primary and metastatic brain tumors. Front. Neurol. 2020, 11, 544680. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Snyder, M.P. Integrative omics for health and disease. Nat. Rev. Genet. 2018, 19, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Bustos, M.A.; Rahimzadeh, N.; Ryu, S.; Gross, R.; Tran, L.T.; Renteria-Lopez, V.M.; Ramos, R.I.; Eisenberg, A.; Hothi, P.; Kesari, S. Cell-free plasma microRNAs that identify patients with glioblastoma. Lab. Investig. 2022, 102, 711–721. [Google Scholar] [CrossRef]
- Bauman, M.M.; Bouchal, S.M.; Monie, D.D.; Aibaidula, A.; Singh, R.; Parney, I.F. Strategies, considerations, and recent advancements in the development of liquid biopsy for glioblastoma: A step towards individualized medicine in glioblastoma. Neurosurg. Focus 2022, 53, E14. [Google Scholar]
- Kopkova, A.; Sana, J.; Fadrus, P.; Slaby, O. Cerebrospinal fluid microRNAs as diagnostic biomarkers in brain tumors. Clin. Chem. Lab. Med. (CCLM) 2018, 56, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.; Warmoes, M.O.; Shen, X.; Locasale, J.W. Epigenetics and cancer metabolism. Cancer Lett. 2015, 356, 309–314. [Google Scholar] [CrossRef] [Green Version]
- Dong, Z.; Cui, H. Epigenetic modulation of metabolism in glioblastoma. Semin. Cancer Biol. 2019, 57, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Senhaji, N.; Squalli Houssaini, A.; Lamrabet, S.; Louati, S.; Bennis, S. Molecular and circulating biomarkers in patients with glioblastoma. Int. J. Mol. Sci. 2022, 23, 7474. [Google Scholar] [CrossRef] [PubMed]
- Stichel, D.; Ebrahimi, A.; Reuss, D.; Schrimpf, D.; Ono, T.; Shirahata, M.; Reifenberger, G.; Weller, M.; Hänggi, D.; Wick, W. Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and TERT promoter mutation in brain tumors and their potential for the reclassification of IDH wt astrocytoma to glioblastoma. Acta Neuropathol. 2018, 136, 793–803. [Google Scholar] [CrossRef] [Green Version]
- Ortiz-Quintero, B. Cell-free microRNAs in blood and other body fluids, as cancer biomarkers. Cell Prolif. 2016, 49, 281–303. [Google Scholar] [CrossRef] [Green Version]
- Zanganeh, S.; Goodarzi, N.; Doroudian, M.; Movahed, E. Potential COVID-19 therapeutic approaches targeting angiotensin-converting enzyme 2; an updated review. Rev. Med. Virol. 2022, 32, e2321. [Google Scholar] [CrossRef]
- Swellam, M.; Ezz El Arab, L.; Al-Posttany, A.S.; Said, S. Clinical impact of circulating oncogenic MiRNA-221 and MiRNA-222 in glioblastoma multiform. J. Neurooncol. 2019, 144, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Garcia, C.M.; Toms, S.A. The role of circulating microRNA in glioblastoma liquid biopsy. World Neurosurg. 2020, 138, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Skog, J.; Würdinger, T.; Van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Yang, C.H.; Yue, J.; Pfeffer, S.R.; Fan, M.; Paulus, E.; Hosni-Ahmed, A.; Sims, M.; Qayyum, S.; Davidoff, A.M.; Handorf, C.R. MicroRNA-21 promotes glioblastoma tumorigenesis by down-regulating insulin-like growth factor-binding protein-3 (IGFBP3). J. Biol. Chem. 2014, 289, 25079–25087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilhan-Mutlu, A.; Wagner, L.; Wöhrer, A.; Furtner, J.; Widhalm, G.; Marosi, C.; Preusser, M. Plasma MicroRNA-21 concentration may be a useful biomarker in glioblastoma patients. Cancer Investig. 2012, 30, 615–621. [Google Scholar] [CrossRef]
- Li, W.; Guo, F.; Wang, P.; Hong, S.; Zhang, C. miR-221/222 confers radioresistance in glioblastoma cells through activating Akt independent of PTEN status. Curr. Mol. Med. 2014, 14, 185–195. [Google Scholar] [CrossRef]
- Zhao, H.; Shen, J.; Hodges, T.R.; Song, R.; Fuller, G.N.; Heimberger, A.B. Serum microRNA profiling in patients with glioblastoma: A survival analysis. Mol. Cancer 2017, 16, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Herman, A.; Gruden, K.; Blejec, A.; Podpečan, V.; Motaln, H.; Rožman, P.; Hren, M.; Zupančič, K.; Veber, M.; Verbovšek, U. Analysis of glioblastoma patients’ plasma revealed the presence of microRNAs with a prognostic impact on survival and those of viral origin. PLoS ONE 2015, 10, e0125791. [Google Scholar] [CrossRef]
- Dong, L.; Li, Y.; Han, C.; Wang, X.; She, L.; Zhang, H. miRNA microarray reveals specific expression in the peripheral blood of glioblastoma patients. Int. J. Oncol. 2014, 45, 746–756. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Ta, W.-W.; Sun, P.-F.; Meng, Y.-F.; Zhao, C.-Z. Diagnostic and prognostic significance of serum miR-145-5p expression in glioblastoma. Int. J. Clin. Exp. Pathol. 2019, 12, 2536. [Google Scholar]
- Regazzo, G.; Terrenato, I.; Spagnuolo, M.; Carosi, M.; Cognetti, G.; Cicchillitti, L.; Sperati, F.; Villani, V.; Carapella, C.; Piaggio, G. A restricted signature of serum miRNAs distinguishes glioblastoma from lower grade gliomas. J. Exp. Clin. Cancer Res. 2016, 35, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Li, Y.; Li, Y.; Shi, X.; Chen, H. Circulating microRNA-137 is a potential biomarker for human glioblastoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3599–3604. [Google Scholar] [PubMed]
- Chen, J.; Yang, L.; Wang, X. Reduced circulating microRNA-203 predicts poor prognosis for glioblastoma. Cancer Biomark. 2017, 20, 521–526. [Google Scholar] [CrossRef]
- Roth, P.; Wischhusen, J.; Happold, C.; Chandran, P.A.; Hofer, S.; Eisele, G.; Weller, M.; Keller, A. A specific miRNA signature in the peripheral blood of glioblastoma patients. J. Neurochem. 2011, 118, 449–457. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.-Q.; Zhang, M.-Y.; Deng, M.-L.; Weng, N.-Q.; Wang, H.-Y.; Wu, S.-X. Low serum level of miR-485-3p predicts poor survival in patients with glioblastoma. PLoS ONE 2017, 12, e0184969. [Google Scholar]
- Akers, J.C.; Hua, W.; Li, H.; Ramakrishnan, V.; Yang, Z.; Quan, K.; Zhu, W.; Li, J.; Figueroa, J.; Hirshman, B.R. A cerebrospinal fluid microRNA signature as biomarker for glioblastoma. Oncotarget 2017, 8, 68769. [Google Scholar] [CrossRef] [Green Version]
- Teplyuk, N.M.; Mollenhauer, B.; Gabriely, G.; Giese, A.; Kim, E.; Smolsky, M.; Kim, R.Y.; Saria, M.G.; Pastorino, S.; Kesari, S. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol. 2012, 14, 689–700. [Google Scholar] [CrossRef] [Green Version]
- Akers, J.C.; Ramakrishnan, V.; Kim, R.; Skog, J.; Nakano, I.; Pingle, S.; Kalinina, J.; Hua, W.; Kesari, S.; Mao, Y. MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): A platform for glioblastoma biomarker development. PLoS ONE 2013, 8, e78115. [Google Scholar]
- Zan, X.-Y.; Li, L. Construction of lncRNA-mediated ceRNA network to reveal clinically relevant lncRNA biomarkers in glioblastomas. Oncol. Lett. 2019, 17, 4369–4374. [Google Scholar] [CrossRef] [Green Version]
- Ita, M.I.; Wang, J.H.; Toulouse, A.; Lim, C.; Fanning, N.; O’Sullivan, M.; Nolan, Y.; Kaar, G.F.; Redmond, H.P. The utility of plasma circulating cell-free messenger RNA as a biomarker of glioma: A pilot study. Acta Neurochir. 2022, 164, 723–735. [Google Scholar]
- Cilibrasi, C.; Simon, T.; Vintu, M.; Tolias, C.; Samuels, M.; Mazarakis, N.K.; Eravci, M.; Stewart, N.; Critchley, G.; Giamas, G. Definition of an inflammatory biomarker signature in plasma-derived extracellular vesicles of glioblastoma patients. Biomedicines 2022, 10, 125. [Google Scholar] [CrossRef]
- Nahand, J.S.; Vandchali, N.R.; Darabi, H.; Doroudian, M.; Banafshe, H.R.; Moghoofei, M.; Babaei, F.; Salmaninejad, A.; Mirzaei, H. Exosomal microRNAs: Novel players in cervical cancer. Epigenomics 2020, 12, 1651–1660. [Google Scholar] [CrossRef]
- Azhdari, M.H.; Goodarzi, N.; Doroudian, M.; MacLoughlin, R. Molecular insight into the therapeutic effects of stem cell-derived exosomes in respiratory diseases and the potential for pulmonary delivery. Int. J. Mol. Sci. 2022, 23, 6273. [Google Scholar] [CrossRef]
- Yekula, A.; Muralidharan, K.; Kang, K.M.; Wang, L.; Balaj, L.; Carter, B.S. From laboratory to clinic: Translation of extracellular vesicle based cancer biomarkers. Methods 2020, 177, 58–66. [Google Scholar] [CrossRef]
- Whitehead, C.A.; Kaye, A.H.; Drummond, K.J.; Widodo, S.S.; Mantamadiotis, T.; Vella, L.J.; Stylli, S.S. Extracellular vesicles and their role in glioblastoma. Crit. Rev. Clin. Lab. Sci. 2020, 57, 227–252. [Google Scholar] [CrossRef] [PubMed]
- Tzaridis, T.; Weller, J.; Bachurski, D.; Shakeri, F.; Schaub, C.; Hau, P.; Buness, A.; Schlegel, U.; Steinbach, J.P.; Seidel, C. A novel serum extracellular vesicle protein signature to monitor glioblastoma tumor progression. Int. J. Cancer 2023, 152, 308–319. [Google Scholar] [CrossRef] [PubMed]
- Lane, R.; Simon, T.; Vintu, M.; Solkin, B.; Koch, B.; Stewart, N.; Benstead-Hume, G.; Pearl, F.M.; Critchley, G.; Stebbing, J. Cell-derived extracellular vesicles can be used as a biomarker reservoir for glioblastoma tumor subtyping. Commun. Biol. 2019, 2, 315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanjani, N.A.; Esmaelizad, N.; Zanganeh, S.; Gharavi, A.T.; Heidarizadeh, P.; Radfar, M.; Omidi, F.; MacLoughlin, R.; Doroudian, M. Emerging role of exosomes as biomarkers in cancer treatment and diagnosis. Crit. Rev. Oncol. Hematol. 2022, 169, 103565. [Google Scholar] [CrossRef] [PubMed]
- Gharavi, A.T.; Hanjani, N.A.; Movahed, E.; Doroudian, M. The role of macrophage subtypes and exosomes in immunomodulation. Cell. Mol. Biol. Lett. 2022, 27, 83. [Google Scholar] [CrossRef]
- Del Bene, M.; Osti, D.; Faletti, S.; Beznoussenko, G.V.; DiMeco, F.; Pelicci, G. Extracellular vesicles: The key for precision medicine in glioblastoma. Neuro Oncol. 2022, 24, 184–196. [Google Scholar]
- Doroudian, M.; Zanganeh, S.; Abbasgholinejad, E.; Donnelly, S.C. Nanomedicine in Lung Cancer Immunotherapy. Front. Bioeng. Biotechnol. 2023, 11, 1144653. [Google Scholar] [CrossRef]
- Russo, M.N.; Whaley, L.A.; Norton, E.S.; Zarco, N.; Guerrero-Cázares, H. Extracellular vesicles in the glioblastoma microenvironment: A diagnostic and therapeutic perspective. Mol. Asp. Med. 2022, 91, 101167. [Google Scholar]
- Sleeman, J.; Moll, J.; Sherman, L.; Dall, P.; Pals, S.T.; Ponta, H.; Herrlich, P. The role of CD44 splice variants in human metastatic cancer. In Ciba Foundation Symposium 189-Cell Adhesion and Human Disease: Cell Adhesion and Human Disease: Ciba Foundation Symposium 189; John Wiley & Sons: Hoboken, NJ, USA, 2007; pp. 142–156. [Google Scholar]
- Ghasempour, E.; Hesami, S.; Movahed, E.; Keshel, S.H.; Doroudian, M. Mesenchymal stem cell-derived exosomes as a new therapeutic strategy in the brain tumors. Stem Cell. Res. Ther. 2022, 13, 527. [Google Scholar] [CrossRef]
- Mukherjee, S.; Pillai, P.P. Current insights on extracellular vesicle-mediated glioblastoma progression: Implications in drug resistance and epithelial-mesenchymal transition. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2022, 1866, 130065. [Google Scholar]
- Cumba Garcia, L.M.; Bouchal, S.M.; Bauman, M.M.; Parney, I.F. Advancements and Technical Considerations for Extracellular Vesicle Isolation and Biomarker Identification in Glioblastoma. Neurosurgery 2023, 93, 33–42. [Google Scholar] [CrossRef]
- Cheng, J.; Meng, J.; Zhu, L.; Peng, Y. Exosomal noncoding RNAs in Glioma: Biological functions and potential clinical applications. Mol. Cancer 2020, 19, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Heydari, R.; Koohi, F.; Rasouli, M.; Rezaei, K.; Abbasgholinejad, E.; Bekeschus, S.; Doroudian, M. Exosomes as Rheumatoid Arthritis Diagnostic Biomarkers and Therapeutic Agents. Vaccines 2023, 11, 687. [Google Scholar] [CrossRef]
- Luo, H.; Zhang, H.; Mao, J.; Cao, H.; Tao, Y.; Zhao, G.; Zhang, Z.; Zhang, N.; Liu, Z.; Zhang, J. Exosome-based nanoimmunotherapy targeting TAMs, a promising strategy for glioma. Cell Death Dis. 2023, 14, 235. [Google Scholar] [CrossRef]
- Vaidya, M.; Sugaya, K. DNA associated with circulating exosomes as a biomarker for glioma. Genes 2020, 11, 1276. [Google Scholar] [CrossRef]
- Naryzhny, S.; Volnitskiy, A.; Kopylov, A.; Zorina, E.; Kamyshinsky, R.; Bairamukov, V.; Garaeva, L.; Shlikht, A.; Shtam, T. Proteome of glioblastoma-derived exosomes as a source of biomarkers. Biomedicines 2020, 8, 216. [Google Scholar] [CrossRef]
- Khatami, S.H.; Karami, N.; Taheri-Anganeh, M.; Taghvimi, S.; Tondro, G.; Khorsand, M.; Soltani Fard, E.; Sedighimehr, N.; Kazemi, M.; Rahimi Jaberi, K. Exosomes: Promising Delivery Tools for Overcoming Blood-Brain Barrier and Glioblastoma Therapy. Mol. Neurobiol. 2023, 60, 4659–4678. [Google Scholar]
- Yang, Q.; Wei, B.; Peng, C.; Wang, L.; Li, C. Identification of serum exosomal miR-98–5p, miR-183–5p, miR-323–3p and miR-19b-3p as potential biomarkers for glioblastoma patients and investigation of their mechanisms. Curr. Res. Transl. Med. 2022, 70, 103315. [Google Scholar] [PubMed]
- Wang, J.; Yue, B.-L.; Huang, Y.-Z.; Lan, X.-Y.; Liu, W.-J.; Chen, H. Exosomal RNAs: Novel potential biomarkers for diseases—A review. Int. J. Mol. Sci. 2022, 23, 2461. [Google Scholar] [PubMed]
- Shao, H.; Chung, J.; Lee, K.; Balaj, L.; Min, C.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Lee, H.; Weissleder, R. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat. Commun. 2015, 6, 6999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Wang, F.; Wang, K.; Zhong, Y.; Wei, X.; Wang, Q.; Zhang, H. Engineered exosomes: A promising drug delivery strategy for brain diseases. Curr. Med. Chem. 2022, 29, 3111–3124. [Google Scholar]
- Wang, L.; Wang, D.; Ye, Z.; Xu, J. Engineering Extracellular Vesicles as Delivery Systems in Therapeutic Applications. Adv. Sci. 2023, 10, 2300552. [Google Scholar] [CrossRef]
- Kar, R.; Dhar, R.; Mukherjee, S.; Nag, S.; Gorai, S.; Mukerjee, N.; Mukherjee, D.; Vatsa, R.; Chandrakanth Jadhav, M.; Ghosh, A. Exosome-based smart drug delivery tool for cancer theranostics. ACS Biomater. Sci. Eng. 2023, 9, 577–594. [Google Scholar] [CrossRef]
- Bian, X.; Xiao, Y.-T.; Wu, T.; Yao, M.; Du, L.; Ren, S.; Wang, J. Microvesicles and chemokines in tumor microenvironment: Mediators of intercellular communications in tumor progression. Mol. Cancer 2019, 18, 50. [Google Scholar] [CrossRef]
- Mahmoudi, K.; Ezrin, A.; Hadjipanayis, C. Small extracellular vesicles as tumor biomarkers for glioblastoma. Mol. Asp. Med. 2015, 45, 97–102. [Google Scholar] [CrossRef]
- Rackles, E.; Lopez, P.H.; Falcon-Perez, J.M. Extracellular vesicles as source for the identification of minimally invasive molecular signatures in glioblastoma. Semin. Cancer Biol. 2022, 87, 148–159. [Google Scholar] [CrossRef]
- Menck, K.; Sivaloganathan, S.; Bleckmann, A.; Binder, C. Microvesicles in cancer: Small size, large potential. Int. J. Mol. Sci. 2020, 21, 5373. [Google Scholar] [CrossRef] [PubMed]
- Simionescu, N.; Nemecz, M.; Petrovici, A.-R.; Nechifor, I.S.; Buga, R.-C.; Dabija, M.G.; Eva, L.; Georgescu, A. Microvesicles and Microvesicle-Associated microRNAs Reflect Glioblastoma Regression: Microvesicle-Associated miR-625-5p Has Biomarker Potential. Int. J. Mol. Sci. 2022, 23, 8398. [Google Scholar] [CrossRef] [PubMed]


| Study Title | Status | Biospecimen | Study Type and Time Frame | ClinicalTrials.gov Identifier |
|---|---|---|---|---|
| Unique Blood and CSF Metabolic Profile Association with Gliomas in Adults | Recruiting |
|
| NCT03865355 |
| A Biospecimen Collection Study in BRAF-V600E Mutated Recurrent Gliomas | Recruiting |
|
| NCT03593993 |
| Testing Cerebrospinal Fluid for Cell-free Tumor DNA in Children, Adolescents, and Young Adults with Brain Tumors | Not Yet Recruiting |
|
| NCT05934630 |
| Evaluating the Role of Cerebrospinal Fluid (CSF) Cell-free DNA (cfDNA) as a Prognostic Biomarker in Glioblastoma | Recruiting |
|
| NCT05927610 |
| Longitudinal Assessment of Marrow and Blood in Patients with Glioblastoma | Not Yet Recruiting |
|
| NCT04657146 |
| Sonobiopsy for Noninvasive and Sensitive Detection of Glioblastoma | Recruiting |
|
| NCT05281731 |
| Profiling Program of Cancer Patients with Sequential Tumor and Liquid Biopsies | Recruiting |
|
| NCT05099068 |
| Evaluating the Expression Levels of MicroRNA-10b in Patients with Gliomas | Recruiting |
|
| NCT01849952 |
| MiRNA | Body Fluid | Regulatory Status | References |
|---|---|---|---|
| miR-21 | Blood | Upregulated | [21,63,64,65] |
| miR-221/222 | Blood | Upregulated | [66] |
| miR-106a-5p, miR-20a-5p, miR-222-3p | Blood | Upregulated | [67] |
| miR-514a-3p, miR-592 | Blood | Upregulated | [68] |
| miR-340, miR-626, miR-576-5p | Blood | Upregulated | [69] |
| miR-145-5p | Blood | Downregulated | [70] |
| miR-125b | Blood | Downregulated | [71] |
| miR-137 | Blood | Downregulated | [72] |
| miR-203 | Blood | Downregulated | [73] |
| miR-342-3p, miR-628-3p | Blood | Downregulated | [74] |
| miR-485-3p | Blood | Downregulated | [75] |
| miR-128 | Blood and Plasma | Downregulated in plasma Upregulated in whole blood | [74] |
| miR-21, miR-10b | CSF | Upregulated | [76,77] |
| miR-21, miR-15b | CSF | Upregulated | [77] |
| miR-17, miR-25, miR-23a, miR-27a, miR-106, miR-130 | CSF | Upregulated | [77] |
| miR-193b-3p, miR-331-3p, miR-21-5p, miR-218-5p, miR374a-5p | CSF | Upregulated | [76] |
| Exosomal miR-21 | CSF | Upregulated | [78] |
| miR-128 | CSF | Downregulated | [77] |
| miR27b-3p, miR-30b-3p, miR548c-3p, miR520f-3p | CSF | Downregulated | [76] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zanganeh, S.; Abbasgholinejad, E.; Doroudian, M.; Esmaelizad, N.; Farjadian, F.; Benhabbour, S.R. The Current Landscape of Glioblastoma Biomarkers in Body Fluids. Cancers 2023, 15, 3804. https://doi.org/10.3390/cancers15153804
Zanganeh S, Abbasgholinejad E, Doroudian M, Esmaelizad N, Farjadian F, Benhabbour SR. The Current Landscape of Glioblastoma Biomarkers in Body Fluids. Cancers. 2023; 15(15):3804. https://doi.org/10.3390/cancers15153804
Chicago/Turabian StyleZanganeh, Saba, Elham Abbasgholinejad, Mohammad Doroudian, Nazanin Esmaelizad, Fatemeh Farjadian, and Soumya Rahima Benhabbour. 2023. "The Current Landscape of Glioblastoma Biomarkers in Body Fluids" Cancers 15, no. 15: 3804. https://doi.org/10.3390/cancers15153804