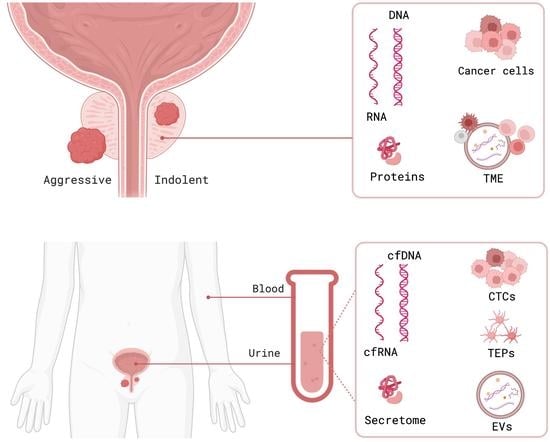
Biomarkers for the Detection and Risk Stratification of Aggressive Prostate Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Tissue Prostate Cancer Biomarkers
2.1. Genomic and Epigenomic Tissue Biomarkers
2.1.1. DNA Mutations and Copy Number Alterations
2.1.2. DNA Methylation
2.1.3. Histone Modification
2.1.4. RNA
2.2. Gene Expression and Proteomic Biomarkers
3. Biomarkers in the Tumor Microenvironment
4. Liquid Biopsy Biomarkers in Prostate Cancer
4.1. Cell-Free DNA
4.1.1. Total cfDNA Amount and Integrity
4.1.2. Genomic Analysis of cfDNA
4.1.3. cfDNA Methylation
4.2. Cell-Free RNA
4.3. Extracellular Vesicles
4.4. Circulating Tumor Cells and Secretome
4.5. Tumor-Educated Platelets
5. Importance of Bioinformatic Tools in Prostate Cancer Detection and Risk Stratification
6. Multi-Parametric Methods for Identifying Aggressive Prostate Cancer
6.1. Integration of Different Diagnostic/Prognostic Approaches
6.2. Multi-Parametric Approach within Liquid Biopsy Profiles
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Gandaglia, G.; Leni, R.; Bray, F.; Fleshner, N.; Freedland, S.J.; Kibel, A.; Stattin, P.; Van Poppel, H.; La Vecchia, C. Epidemiology and Prevention of Prostate Cancer. Eur. Urol. Oncol. 2021, 4, 877–892. [Google Scholar] [CrossRef] [PubMed]
- Rebello, R.J.; Oing, C.; Knudsen, K.E.; Loeb, S.; Johnson, D.C.; Reiter, R.E.; Gillessen, S.; Van der Kwast, T.; Bristow, R.G. Prostate cancer. Nat. Rev. Dis. Primers 2021, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.; Smith, S.; Shamash, J. Metastatic Hormone-Sensitive Prostate Cancer (mHSPC): Advances and Treatment Strategies in the First-Line Setting. Oncol. Ther. 2020, 8, 209–230. [Google Scholar] [CrossRef] [PubMed]
- Sartor, O.; de Bono, J.S. Metastatic Prostate Cancer. N. Engl. J. Med. 2018, 378, 645–657. [Google Scholar] [CrossRef]
- Van Poppel, H.; Albreht, T.; Basu, P.; Hogenhout, R.; Collen, S.; Roobol, M. Serum PSA-based early detection of prostate cancer in Europe and globally: Past, present and future. Nat. Rev. Urol. 2022, 19, 562–572. [Google Scholar] [CrossRef]
- Schröder, F.H.; Hugosson, J.; Carlsson, S.; Tammela, T.; Määttänen, L.; Auvinen, A.; Kwiatkowski, M.; Recker, F.; Roobol, M.J. Screening for prostate cancer decreases the risk of developing metastatic disease: Findings from the European Randomized Study of Screening for Prostate Cancer (ERSPC). Eur. Urol. 2012, 62, 745–752. [Google Scholar] [CrossRef]
- Schröder, F.H.; Hugosson, J.; Roobol, M.J.; Tammela, T.L.; Ciatto, S.; Nelen, V.; Kwiatkowski, M.; Lujan, M.; Lilja, H.; Zappa, M.; et al. Prostate-cancer mortality at 11 years of follow-up. N. Engl. J. Med. 2012, 366, 981–990. [Google Scholar] [CrossRef]
- Pierre-Victor, D.; Parnes, H.L.; Andriole, G.L.; Pinsky, P.F. Prostate Cancer Incidence and Mortality Following a Negative Biopsy in a Population Undergoing PSA Screening. Urology 2021, 155, 62–69. [Google Scholar] [CrossRef]
- Saini, S. PSA and beyond: Alternative prostate cancer biomarkers. Cell. Oncol. 2016, 39, 97–106. [Google Scholar] [CrossRef]
- Loeb, S.; Bjurlin, M.A.; Nicholson, J.; Tammela, T.L.; Penson, D.F.; Carter, H.B.; Carroll, P.; Etzioni, R. Overdiagnosis and overtreatment of prostate cancer. Eur. Urol. 2014, 65, 1046–1055. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Wang, Z.Q.; Li, M.; Zhou, M.Y.; Yu, Y.F.; Zhan, W.W. Establishment of two new predictive models for prostate cancer to determine whether to require prostate biopsy when the PSA level is in the diagnostic gray zone (4–10 ng mL−1). Asian J. Androl. 2020, 22, 213–216. [Google Scholar] [CrossRef] [PubMed]
- Clinton, T.N.; Bagrodia, A.; Lotan, Y.; Margulis, V.; Raj, G.V.; Woldu, S.L. Tissue-based biomarkers in prostate cancer. Expert Rev. Precis. Med. Drug Dev. 2017, 2, 249–260. [Google Scholar] [CrossRef] [Green Version]
- Massie, C.E.; Mills, I.G.; Lynch, A.G. The importance of DNA methylation in prostate cancer development. J. Steroid Biochem. Mol. Biol. 2017, 166, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomlins, S.A.; Rhodes, D.R.; Perner, S.; Dhanasekaran, S.M.; Mehra, R.; Sun, X.W.; Varambally, S.; Cao, X.; Tchinda, J.; Kuefer, R.; et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005, 310, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Van Neste, L.; Herman, J.G.; Otto, G.; Bigley, J.W.; Epstein, J.I.; Van Criekinge, W. The epigenetic promise for prostate cancer diagnosis. Prostate 2012, 72, 1248–1261. [Google Scholar] [CrossRef]
- Robinson, D.; Van Allen, E.M.; Wu, Y.-M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.-M.; Montgomery, B.; Taplin, M.-E.; Pritchard, C.C.; Attard, G.; et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef] [Green Version]
- Haffner, M.C.; Zwart, W.; Roudier, M.P.; True, L.D.; Nelson, W.G.; Epstein, J.I.; De Marzo, A.M.; Nelson, P.S.; Yegnasubramanian, S. Genomic and phenotypic heterogeneity in prostate cancer. Nat. Rev. Urol. 2021, 18, 79–92. [Google Scholar] [CrossRef]
- Castro, E.; Goh, C.; Olmos, D.; Saunders, E.; Leongamornlert, D.; Tymrakiewicz, M.; Mahmud, N.; Dadaev, T.; Govindasami, K.; Guy, M.; et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J. Clin. Oncol. 2013, 31, 1748–1757. [Google Scholar] [CrossRef] [Green Version]
- Attard, G.; Parker, C.; Eeles, R.A.; Schröder, F.; Tomlins, S.A.; Tannock, I.; Drake, C.G.; de Bono, J.S. Prostate cancer. Lancet 2016, 387, 70–82. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Mateo, J.; Walsh, M.F.; De Sarkar, N.; Abida, W.; Beltran, H.; Garofalo, A.; Gulati, R.; Carreira, S.; Eeles, R.; et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N. Engl. J. Med. 2016, 375, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.; Alkhushaym, N.; Fallatah, S.; Althagafi, A.; Aljadeed, R.; Alsowaida, Y.; Jeter, J.; Martin, J.R.; Babiker, H.M.; McBride, A.; et al. The association of BRCA1 and BRCA2 mutations with prostate cancer risk, frequency, and mortality: A meta-analysis. Prostate 2019, 79, 880–895. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; Fisher, C.; Foster, C.S.; Jameson, C.; Barbachanno, Y.; Bartlett, J.; Bancroft, E.; Doherty, R.; Kote-Jarai, Z.; Peock, S.; et al. Prostate cancer in male BRCA1 and BRCA2 mutation carriers has a more aggressive phenotype. Br. J. Cancer 2008, 98, 502–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dall’Era, M.A.; McPherson, J.D.; Gao, A.C.; DeVere White, R.W.; Gregg, J.P.; Lara, P.N., Jr. Germline and somatic DNA repair gene alterations in prostate cancer. Cancer 2020, 126, 2980–2985. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015, 163, 1011–1025. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, G.; Armenia, J.; Mazzu, Y.Z.; Nandakumar, S.; Stopsack, K.H.; Atiq, M.O.; Komura, K.; Jehane, L.; Hirani, R.; Chadalavada, K.; et al. Significance of BRCA2 and RB1 Co-loss in Aggressive Prostate Cancer Progression. Clin. Cancer Res. 2020, 26, 2047–2064. [Google Scholar] [CrossRef]
- Beltran, H.; Yelensky, R.; Frampton, G.M.; Park, K.; Downing, S.R.; MacDonald, T.Y.; Jarosz, M.; Lipson, D.; Tagawa, S.T.; Nanus, D.M.; et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur. Urol. 2013, 63, 920–926. [Google Scholar] [CrossRef] [Green Version]
- Hamid, A.A.; Gray, K.P.; Shaw, G.; MacConaill, L.E.; Evan, C.; Bernard, B.; Loda, M.; Corcoran, N.M.; Van Allen, E.M.; Choudhury, A.D.; et al. Compound Genomic Alterations of TP53, PTEN, and RB1 Tumor Suppressors in Localized and Metastatic Prostate Cancer. Eur. Urol. 2019, 76, 89–97. [Google Scholar] [CrossRef]
- Arora, K.; Barbieri, C.E. Molecular Subtypes of Prostate Cancer. Curr. Oncol. Rep. 2018, 20, 58. [Google Scholar] [CrossRef]
- Deek, M.P.; Van der Eecken, K.; Phillips, R.; Parikh, N.R.; Isaacsson Velho, P.; Lotan, T.L.; Kishan, A.U.; Maurer, T.; Boutros, P.C.; Hovens, C.; et al. The Mutational Landscape of Metastatic Castration-sensitive Prostate Cancer: The Spectrum Theory Revisited. Eur. Urol. 2021, 80, 632–640. [Google Scholar] [CrossRef]
- Teroerde, M.; Nientiedt, C.; Duensing, A.; Hohenfellner, M.; Stenzinger, A.; Duensing, S. Revisiting the Role of p53 in Prostate Cancer. In Prostate Cancer; Bott, S.R.J., Ng, K.L., Eds.; Exon Publications: Brisbane, Australia, 2021. [Google Scholar] [CrossRef]
- Nientiedt, C.; Budczies, J.; Endris, V.; Kirchner, M.; Schwab, C.; Jurcic, C.; Behnisch, R.; Hoveida, S.; Lantwin, P.; Kaczorowski, A.; et al. Mutations in TP53 or DNA damage repair genes define poor prognostic subgroups in primary prostate cancer. Urol. Oncol. 2022, 40, 8.e11–18.e18. [Google Scholar] [CrossRef]
- Aggarwal, R.R.; Quigley, D.A.; Huang, J.; Zhang, L.; Beer, T.M.; Rettig, M.B.; Reiter, R.E.; Gleave, M.E.; Thomas, G.V.; Foye, A.; et al. Whole-Genome and Transcriptional Analysis of Treatment-Emergent Small-Cell Neuroendocrine Prostate Cancer Demonstrates Intraclass Heterogeneity. Mol. Cancer Res. 2019, 17, 1235–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abida, W.; Cyrta, J.; Heller, G.; Prandi, D.; Armenia, J.; Coleman, I.; Cieslik, M.; Benelli, M.; Robinson, D.; Van Allen, E.M.; et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 11428–11436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangolini, A.; Rocca, C.; Bassi, C.; Ippolito, C.; Negrini, M.; Dell’Atti, L.; Lanza, G.; Gafà, R.; Bianchi, N.; Pinton, P.; et al. Detection of disease-causing mutations in prostate cancer by NGS sequencing. Cell Biol. Int. 2022, 46, 1047–1061. [Google Scholar] [CrossRef]
- Taylor, B.S.; Schultz, N.; Hieronymus, H.; Gopalan, A.; Xiao, Y.; Carver, B.S.; Arora, V.K.; Kaushik, P.; Cerami, E.; Reva, B.; et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010, 18, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Quigley, D.A.; Dang, H.X.; Zhao, S.G.; Lloyd, P.; Aggarwal, R.; Alumkal, J.J.; Foye, A.; Kothari, V.; Perry, M.D.; Bailey, A.M.; et al. Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell 2018, 174, 758–769.e759. [Google Scholar] [CrossRef] [Green Version]
- Boysen, G.; Barbieri, C.E.; Prandi, D.; Blattner, M.; Chae, S.S.; Dahija, A.; Nataraj, S.; Huang, D.; Marotz, C.; Xu, L.; et al. SPOP mutation leads to genomic instability in prostate cancer. eLife 2015, 4, e09207. [Google Scholar] [CrossRef]
- Cotter, K.; Rubin, M.A. The evolving landscape of prostate cancer somatic mutations. Prostate 2022, 82 (Suppl. 1), S13–S24. [Google Scholar] [CrossRef]
- Sooreshjani, M.A.; Nikhil, K.; Kamra, M.; Nguyen, D.N.; Kumar, D.; Shah, K. LIMK2-NKX3.1 Engagement Promotes Castration-Resistant Prostate Cancer. Cancers 2021, 13, 2324. [Google Scholar] [CrossRef]
- Asatiani, E.; Huang, W.X.; Wang, A.; Rodriguez Ortner, E.; Cavalli, L.R.; Haddad, B.R.; Gelmann, E.P. Deletion, methylation, and expression of the NKX3.1 suppressor gene in primary human prostate cancer. Cancer Res. 2005, 65, 1164–1173. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.Y.; Ding, L.; Zhou, T.R.; Yan, T.; Li, J.; Liang, C. FOXA1 in prostate cancer. Asian J. Androl. 2022. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Zhang, J.; Hu, Q.; Zhi, F.; Zhang, S.; Mao, D.; Zhang, Y.; Liang, H. Significance of the TMPRSS2:ERG gene fusion in prostate cancer. Mol. Med. Rep. 2017, 16, 5450–5458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burdova, A.; Bouchal, J.; Tavandzis, S.; Kolar, Z. TMPRSS2-ERG gene fusion in prostate cancer. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2014, 158, 502–510. [Google Scholar] [CrossRef] [Green Version]
- Rosen, P.; Sesterhenn, I.A.; Brassell, S.A.; McLeod, D.G.; Srivastava, S.; Dobi, A. Clinical potential of the ERG oncoprotein in prostate cancer. Nat. Rev. Urol. 2012, 9, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.B.; Bentley, J.; Jeffery, Z.; DeMarzo, A.M. Heterogeneity of PTEN and ERG expression in prostate cancer on core needle biopsies: Implications for cancer risk stratification and biomarker sampling. Hum. Pathol. 2015, 46, 698–706. [Google Scholar] [CrossRef]
- Liu, W.; Xie, C.C.; Thomas, C.Y.; Kim, S.T.; Lindberg, J.; Egevad, L.; Wang, Z.; Zhang, Z.; Sun, J.; Sun, J.; et al. Genetic markers associated with early cancer-specific mortality following prostatectomy. Cancer 2013, 119, 2405–2412. [Google Scholar] [CrossRef] [Green Version]
- Tapia-Laliena, M.A.; Korzeniewski, N.; Hohenfellner, M.; Duensing, S. High-risk prostate cancer: A disease of genomic instability. Urol. Oncol. 2014, 32, 1101–1107. [Google Scholar] [CrossRef]
- Silva, M.P.; Barros-Silva, J.D.; Vieira, J.; Lisboa, S.; Torres, L.; Correia, C.; Vieira-Coimbra, M.; Martins, A.T.; Jerónimo, C.; Henrique, R.; et al. NCOA2 is a candidate target gene of 8q gain associated with clinically aggressive prostate cancer. Genes Chromosomes Cancer 2016, 55, 365–374. [Google Scholar] [CrossRef]
- Faisal, F.A.; Murali, S.; Kaur, H.; Vidotto, T.; Guedes, L.B.; Salles, D.C.; Kothari, V.; Tosoian, J.J.; Han, S.; Hovelson, D.H.; et al. CDKN1B Deletions are Associated with Metastasis in African American Men with Clinically Localized, Surgically Treated Prostate Cancer. Clin. Cancer Res. 2020, 26, 2595–2602. [Google Scholar] [CrossRef]
- Chang, B.L.; Zheng, S.L.; Isaacs, S.D.; Wiley, K.E.; Turner, A.; Li, G.; Walsh, P.C.; Meyers, D.A.; Isaacs, W.B.; Xu, J. A polymorphism in the CDKN1B gene is associated with increased risk of hereditary prostate cancer. Cancer Res. 2004, 64, 1997–1999. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.S.; Je, E.M.; Yoo, N.J.; Lee, S.H. Loss of ARID1A expression is uncommon in gastric, colorectal, and prostate cancers. Apmis 2012, 120, 1020–1022. [Google Scholar] [CrossRef] [PubMed]
- Santric, V.; Djokic, M.; Suvakov, S.; Pljesa-Ercegovac, M.; Nikitovic, M.; Radic, T.; Acimovic, M.; Stankovic, V.; Bumbasirevic, U.; Milojevic, B.; et al. GSTP1 rs1138272 Polymorphism Affects Prostate Cancer Risk. Medicina 2020, 56, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hopkins, J.F.; Sabelnykova, V.Y.; Weischenfeldt, J.; Simon, R.; Aguiar, J.A.; Alkallas, R.; Heisler, L.E.; Zhang, J.; Watson, J.D.; Chua, M.L.K.; et al. Mitochondrial mutations drive prostate cancer aggression. Nat. Commun. 2017, 8, 656. [Google Scholar] [CrossRef] [PubMed]
- Dathathri, E.; Isebia, K.T.; Abali, F.; Lolkema, M.P.; Martens, J.W.M.; Terstappen, L.; Bansal, R. Liquid Biopsy Based Circulating Biomarkers in Metastatic Prostate Cancer. Front. Oncol. 2022, 12, 863472. [Google Scholar] [CrossRef]
- Wyatt, A.W.; Annala, M.; Aggarwal, R.; Beja, K.; Feng, F.; Youngren, J.; Foye, A.; Lloyd, P.; Nykter, M.; Beer, T.M.; et al. Concordance of Circulating Tumor DNA and Matched Metastatic Tissue Biopsy in Prostate Cancer. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef] [Green Version]
- Dor, Y.; Cedar, H. Principles of DNA methylation and their implications for biology and medicine. Lancet 2018, 392, 777–786. [Google Scholar] [CrossRef]
- Jones, P.A.; Baylin, S.B. The epigenomics of cancer. Cell 2007, 128, 683–692. [Google Scholar] [CrossRef] [Green Version]
- Fiano, V.; Zugna, D.; Grasso, C.; Trevisan, M.; Delsedime, L.; Molinaro, L.; Cassoni, P.; Papotti, M.; Merletti, F.; Akre, O.; et al. DNA methylation in repeat negative prostate biopsies as a marker of missed prostate cancer. Clin. Epigenetics 2019, 11, 152. [Google Scholar] [CrossRef]
- Partin, A.W.; Van Neste, L.; Klein, E.A.; Marks, L.S.; Gee, J.R.; Troyer, D.A.; Rieger-Christ, K.; Jones, J.S.; Magi-Galluzzi, C.; Mangold, L.A.; et al. Clinical validation of an epigenetic assay to predict negative histopathological results in repeat prostate biopsies. J. Urol. 2014, 192, 1081–1087. [Google Scholar] [CrossRef] [Green Version]
- Rosenbaum, E.; Hoque, M.O.; Cohen, Y.; Zahurak, M.; Eisenberger, M.A.; Epstein, J.I.; Partin, A.W.; Sidransky, D. Promoter Hypermethylation as an Independent Prognostic Factor for Relapse in Patients with Prostate Cancer Following Radical Prostatectomy. Clin. Cancer Res. 2005, 11, 8321–8325. [Google Scholar] [CrossRef] [Green Version]
- Stewart, G.D.; Van Neste, L.; Delvenne, P.; Delrée, P.; Delga, A.; McNeill, S.A.; O’Donnell, M.; Clark, J.; Van Criekinge, W.; Bigley, J.; et al. Clinical utility of an epigenetic assay to detect occult prostate cancer in histopathologically negative biopsies: Results of the MATLOC study. J. Urol. 2013, 189, 1110–1116. [Google Scholar] [CrossRef] [PubMed]
- Uhl, B.; Gevensleben, H.; Tolkach, Y.; Sailer, V.; Majores, M.; Jung, M.; Meller, S.; Stein, J.; Ellinger, J.; Dietrich, D.; et al. PITX2 DNA Methylation as Biomarker for Individualized Risk Assessment of Prostate Cancer in Core Biopsies. J. Mol. Diagn. 2017, 19, 107–114. [Google Scholar] [PubMed] [Green Version]
- Vasiljević, N.; Ahmad, A.S.; Carter, P.D.; Fisher, G.; Berney, D.M.; Foster, C.S.; Cuzick, J.; Lorincz, A.T. DNA methylation of PITX2 predicts poor survival in men with prostate cancer. Biomark. Med. 2014, 8, 1143–1150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiss, G.; Cottrell, S.; Distler, J.; Schatz, P.; Kristiansen, G.; Ittmann, M.; Haefliger, C.; Lesche, R.; Hartmann, A.; Corman, J.; et al. DNA Methylation of the PITX2 Gene Promoter Region is a Strong Independent Prognostic Marker of Biochemical Recurrence in Patients With Prostate Cancer After Radical Prostatectomy. J. Urol. 2009, 181, 1678–1685. [Google Scholar] [CrossRef]
- Majumdar, S.; Buckles, E.; Estrada, J.; Koochekpour, S. Aberrant DNA methylation and prostate cancer. Curr. Genom. 2011, 12, 486–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMahon, K.W.; Karunasena, E.; Ahuja, N. The Roles of DNA Methylation in the Stages of Cancer. Cancer J. 2017, 23, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Bañez, L.L.; Sun, L.; van Leenders, G.J.; Wheeler, T.M.; Bangma, C.H.; Freedland, S.J.; Ittmann, M.M.; Lark, A.L.; Madden, J.F.; Hartman, A.; et al. Multicenter clinical validation of PITX2 methylation as a prostate specific antigen recurrence predictor in patients with post-radical prostatectomy prostate cancer. J. Urol. 2010, 184, 149–156. [Google Scholar] [CrossRef]
- Daniunaite, K.; Jarmalaite, S.; Kalinauskaite, N.; Petroska, D.; Laurinavicius, A.; Lazutka, J.R.; Jankevicius, F. Prognostic value of RASSF1 promoter methylation in prostate cancer. J. Urol. 2014, 192, 1849–1855. [Google Scholar] [CrossRef]
- Dietrich, D.; Hasinger, O.; Bañez, L.L.; Sun, L.; van Leenders, G.J.; Wheeler, T.M.; Bangma, C.H.; Wernert, N.; Perner, S.; Freedland, S.J.; et al. Development and clinical validation of a real-time PCR assay for PITX2 DNA methylation to predict prostate-specific antigen recurrence in prostate cancer patients following radical prostatectomy. J. Mol. Diagn. 2013, 15, 270–279. [Google Scholar] [CrossRef]
- Maldonado, L.; Brait, M.; Loyo, M.; Sullenberger, L.; Wang, K.; Peskoe, S.B.; Rosenbaum, E.; Howard, R.; Toubaji, A.; Albadine, R.; et al. GSTP1 promoter methylation is associated with recurrence in early stage prostate cancer. J. Urol. 2014, 192, 1542–1548. [Google Scholar] [CrossRef] [Green Version]
- Richiardi, L.; Fiano, V.; Vizzini, L.; De Marco, L.; Delsedime, L.; Akre, O.; Tos, A.G.; Merletti, F. Promoter methylation in APC, RUNX3, and GSTP1 and mortality in prostate cancer patients. J. Clin. Oncol. 2009, 27, 3161–3168. [Google Scholar] [CrossRef] [PubMed]
- Henrique, R.; Ribeiro, F.R.; Fonseca, D.; Hoque, M.O.; Carvalho, A.L.; Costa, V.L.; Pinto, M.; Oliveira, J.; Teixeira, M.R.; Sidransky, D.; et al. High promoter methylation levels of APC predict poor prognosis in sextant biopsies from prostate cancer patients. Clin. Cancer Res. 2007, 13, 6122–6129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Neste, L.; Groskopf, J.; Grizzle, W.E.; Adams, G.W.; DeGuenther, M.S.; Kolettis, P.N.; Bryant, J.E.; Kearney, G.P.; Kearney, M.C.; Van Criekinge, W.; et al. Epigenetic risk score improves prostate cancer risk assessment. Prostate 2017, 77, 1259–1264. [Google Scholar] [CrossRef] [PubMed]
- Van Neste, L.; Partin, A.W.; Stewart, G.D.; Epstein, J.I.; Harrison, D.J.; Van Criekinge, W. Risk score predicts high-grade prostate cancer in DNA-methylation positive, histopathologically negative biopsies. Prostate 2016, 76, 1078–1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, S.; Geybels, M.S.; Leonardson, A.; Rubicz, R.; Kolb, S.; Yan, Q.; Klotzle, B.; Bibikova, M.; Hurtado-Coll, A.; Troyer, D.; et al. Epigenome-Wide Tumor DNA Methylation Profiling Identifies Novel Prognostic Biomarkers of Metastatic-Lethal Progression in Men Diagnosed with Clinically Localized Prostate Cancer. Clin. Cancer Res. 2017, 23, 311–319. [Google Scholar] [CrossRef] [Green Version]
- Mundbjerg, K.; Chopra, S.; Alemozaffar, M.; Duymich, C.; Lakshminarasimhan, R.; Nichols, P.W.; Aron, M.; Siegmund, K.D.; Ukimura, O.; Aron, M.; et al. Identifying aggressive prostate cancer foci using a DNA methylation classifier. Genome Biol. 2017, 18, 3. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Wang, L.; Wang, Q.; Li, W. Histone modifications and chromatin organization in prostate cancer. Epigenomics 2010, 2, 551–560. [Google Scholar] [CrossRef] [Green Version]
- Pang, J.; Yang, Y.W.; Huang, Y.; Yang, J.; Zhang, H.; Chen, R.; Dong, L.; Huang, Y.; Wang, D.; Liu, J.; et al. P110β Inhibition Reduces Histone H3K4 Di-Methylation in Prostate Cancer. Prostate 2017, 77, 299–308. [Google Scholar] [CrossRef]
- Cackowski, F.C.; Heath, E.I. Prostate cancer dormancy and recurrence. Cancer Lett. 2022, 524, 103–108. [Google Scholar] [CrossRef]
- Dryhurst, D.; Ausió, J. Histone H2A.Z deregulation in prostate cancer. Cause or effect? Cancer Metastasis Rev. 2014, 33, 429–439. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Huang, V.; Xu, X.; Livingstone, J.; Soares, F.; Jeon, J.; Zeng, Y.; Hua, J.T.; Petricca, J.; Guo, H.; et al. Widespread and Functional RNA Circularization in Localized Prostate Cancer. Cell 2019, 176, 831–843.e22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.P.; Ding, Y.; Chen, Z.; Liu, S.; Michalopoulos, A.; Chen, R.; Gulzar, Z.G.; Yang, B.; Cieply, K.M.; Luvison, A.; et al. Novel fusion transcripts associate with progressive prostate cancer. Am. J. Pathol. 2014, 184, 2840–2849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, L.B.; Palumbo, A.; de Mello, K.D.; Sternberg, C.; Caetano, M.S.; de Oliveira, F.L.; Neves, A.F.; Nasciutti, L.E.; Goulart, L.R.; Gimba, E.R. PCA3 noncoding RNA is involved in the control of prostate-cancer cell survival and modulates androgen receptor signaling. BMC Cancer 2012, 12, 507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deras, I.L.; Aubin, S.M.; Blase, A.; Day, J.R.; Koo, S.; Partin, A.W.; Ellis, W.J.; Marks, L.S.; Fradet, Y.; Rittenhouse, H.; et al. PCA3: A molecular urine assay for predicting prostate biopsy outcome. J. Urol. 2008, 179, 1587–1592. [Google Scholar] [CrossRef]
- Prensner, J.R.; Zhao, S.; Erho, N.; Schipper, M.; Iyer, M.K.; Dhanasekaran, S.M.; Magi-Galluzzi, C.; Mehra, R.; Sahu, A.; Siddiqui, J.; et al. RNA biomarkers associated with metastatic progression in prostate cancer: A multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 2014, 15, 1469–1480. [Google Scholar] [CrossRef] [Green Version]
- Mehra, R.; Shi, Y.; Udager, A.M.; Prensner, J.R.; Sahu, A.; Iyer, M.K.; Siddiqui, J.; Cao, X.; Wei, J.; Jiang, H.; et al. A novel RNA in situ hybridization assay for the long noncoding RNA SChLAP1 predicts poor clinical outcome after radical prostatectomy in clinically localized prostate cancer. Neoplasia 2014, 16, 1121–1127. [Google Scholar] [CrossRef] [Green Version]
- Cesnik, A.J.; Yang, B.; Truong, A.; Etheridge, T.; Spiniello, M.; Steinbrink, M.I.; Shortreed, M.R.; Frey, B.L.; Jarrard, D.F.; Smith, L.M. Long Noncoding RNAs AC009014.3 and Newly Discovered XPLAID Differentiate Aggressive and Indolent Prostate Cancers. Transl. Oncol. 2018, 11, 808–814. [Google Scholar] [CrossRef]
- Prensner, J.R.; Iyer, M.K.; Balbin, O.A.; Dhanasekaran, S.M.; Cao, Q.; Brenner, J.C.; Laxman, B.; Asangani, I.A.; Grasso, C.S.; Kominsky, H.D.; et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 2011, 29, 742–749. [Google Scholar] [CrossRef]
- Shukla, S.; Zhang, X.; Niknafs, Y.S.; Xiao, L.; Mehra, R.; Cieślik, M.; Ross, A.; Schaeffer, E.; Malik, B.; Guo, S.; et al. Identification and Validation of PCAT14 as Prognostic Biomarker in Prostate Cancer. Neoplasia 2016, 18, 489–499. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rana, S.; Valbuena, G.N.; Curry, E.; Bevan, C.L.; Keun, H.C. MicroRNAs as biomarkers for prostate cancer prognosis: A systematic review and a systematic reanalysis of public data. Br. J. Cancer 2022, 126, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Larne, O.; Martens-Uzunova, E.; Hagman, Z.; Edsjö, A.; Lippolis, G.; den Berg, M.S.; Bjartell, A.; Jenster, G.; Ceder, Y. miQ--a novel microRNA based diagnostic and prognostic tool for prostate cancer. Int. J. Cancer 2013, 132, 2867–2875. [Google Scholar] [CrossRef]
- Kristensen, H.; Thomsen, A.R.; Haldrup, C.; Dyrskjøt, L.; Høyer, S.; Borre, M.; Mouritzen, P.; Ørntoft, T.F.; Sørensen, K.D. Novel diagnostic and prognostic classifiers for prostate cancer identified by genome-wide microRNA profiling. Oncotarget 2016, 7, 30760–30771. [Google Scholar] [CrossRef]
- Feng, S.; Qian, X.; Li, H.; Zhang, X. Combinations of elevated tissue miRNA-17-92 cluster expression and serum prostate-specific antigen as potential diagnostic biomarkers for prostate cancer. Oncol. Lett. 2017, 14, 6943–6949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, L.; Fredsøe, J.; Kristensen, H.; Strand, S.H.; Rasmussen, A.; Høyer, S.; Borre, M.; Mouritzen, P.; Ørntoft, T.; Sørensen, K.D. Training and validation of a novel 4-miRNA ratio model (MiCaP) for prediction of postoperative outcome in prostate cancer patients. Ann. Oncol. 2018, 29, 2003–2009. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhao, H.; Shi, Y.; Yang, F.; Wang, L.T.; Kang, G.; Nie, Y.; Wang, J. Perioperative Dynamic Changes in Circulating Tumor DNA in Patients with Lung Cancer (DYNAMIC). Clin. Cancer Res. 2019, 25, 7058–7067. [Google Scholar] [CrossRef] [Green Version]
- Hansen, E.B.; Fredsøe, J.; Okholm, T.L.H.; Ulhøi, B.P.; Klingenberg, S.; Jensen, J.B.; Kjems, J.; Bouchelouche, K.; Borre, M.; Damgaard, C.K.; et al. The transcriptional landscape and biomarker potential of circular RNAs in prostate cancer. Genome Med. 2022, 14, 8. [Google Scholar] [CrossRef]
- Kappelhoff, R.; Puente, X.S.; Wilson, C.H.; Seth, A.; López-Otín, C.; Overall, C.M. Overview of transcriptomic analysis of all human proteases, non-proteolytic homologs and inhibitors: Organ, tissue and ovarian cancer cell line expression profiling of the human protease degradome by the CLIP-CHIP™ DNA microarray. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 2210–2219. [Google Scholar] [CrossRef]
- Koistinen, H.; Künnapuu, J.; Jeltsch, M. KLK3 in the Regulation of Angiogenesis-Tumorigenic or Not? Int. J. Mol. Sci. 2021, 22, 13545. [Google Scholar] [CrossRef]
- Fuhrman-Luck, R.A.; Loessner, D.; Clements, J.A. Kallikrein-Related Peptidases in Prostate Cancer: From Molecular Function to Clinical Application. Ejifcc 2014, 25, 269–281. [Google Scholar] [PubMed]
- Wu, J.; Guo, S.; Wang, L.; Liao, Z. Correlation analysis between CD133, Klk3 and grhl2 expression and tumor characteristics in prostate cancer. Cell. Mol. Biol. 2022, 67, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Ristau, B.T.; O’Keefe, D.S.; Bacich, D.J. The prostate-specific membrane antigen: Lessons and current clinical implications from 20 years of research. Urol. Oncol. 2014, 32, 272–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Yoon, J.; Ko, D.; Yu, M.; Lee, S.; Kim, S. TMPRSS4 promotes cancer stem–like properties in prostate cancer cells through upregulation of SOX2 by SLUG and TWIST1. J. Exp. Clin. Cancer Res. 2021, 40, 372. [Google Scholar] [CrossRef]
- Santos, N.J.; Camargo, A.C.L.; Carvalho, H.F.; Justulin, L.A.; Felisbino, S.L. Prostate Cancer Secretome and Membrane Proteome from Pten Conditional Knockout Mice Identify Potential Biomarkers for Disease Progression. Int. J. Mol. Sci. 2022, 23, 9224. [Google Scholar] [CrossRef]
- Muller, A.K.; Foll, M.; Heckelmann, B.; Kiefer, S.; Werner, M.; Schilling, O.; Biniossek, M.L.; Jilg, C.A.; Drendel, V. Proteomic Characterization of Prostate Cancer to Distinguish Nonmetastasizing and Metastasizing Primary Tumors and Lymph Node Metastases. Neoplasia 2018, 20, 140–151. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, Y.T.; Chen, Y.; Wang, H.; Young, D.; Shi, T.; Song, Y.; Schepmoes, A.A.; Kuo, C.; Fillmore, T.L.; et al. Proteomic Tissue-Based Classifier for Early Prediction of Prostate Cancer Progression. Cancers 2020, 12, 1268. [Google Scholar] [CrossRef]
- Lygirou, V.; Fasoulakis, K.; Stroggilos, R.; Makridakis, M.; Latosinska, A.; Frantzi, M.; Katafigiotis, I.; Alamanis, C.; Stravodimos, K.G.; Constantinides, C.A.; et al. Proteomic Analysis of Prostate Cancer FFPE Samples Reveals Markers of Disease Progression and Aggressiveness. Cancers 2022, 14, 3765. [Google Scholar] [CrossRef]
- Shipitsin, M.; Small, C.; Choudhury, S.; Giladi, E.; Friedlander, S.; Nardone, J.; Hussain, S.; Hurley, A.D.; Ernst, C.; Huang, Y.E.; et al. Identification of proteomic biomarkers predicting prostate cancer aggressiveness and lethality despite biopsy-sampling error. Br. J. Cancer 2014, 111, 1201–1212. [Google Scholar] [CrossRef]
- Garcia-Marques, F.; Liu, S.; Totten, S.M.; Bermudez, A.; Tanimoto, C.; Hsu, E.C.; Nolley, R.; Hembree, A.; Stoyanova, T.; Brooks, J.D.; et al. Protein signatures to distinguish aggressive from indolent prostate cancer. Prostate 2022, 82, 605–616. [Google Scholar] [CrossRef]
- Ahmad, F.; Cherukuri, M.K.; Choyke, P.L. Metabolic reprogramming in prostate cancer. Br. J. Cancer 2021, 125, 1185–1196. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Zhu, W.; Ding, Y.; Zhu, H.; Jing, X.; Yu, H.; Lu, M.; Qiao, Y.; Wang, X.; Ai, H.; et al. Phosphorylation of LIFR promotes prostate cancer progression by activating the AKT pathway. Cancer Lett. 2019, 451, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Teng, M.; Zhou, S.; Cai, C.; Lupien, M.; He, H.H. Pioneer of prostate cancer: Past, present and the future of FOXA1. Protein Cell 2021, 12, 29–38. [Google Scholar] [CrossRef]
- Josson, S.; Chung, L.W.; Gururajan, M. microRNAs and Prostate Cancer. Adv. Exp. Med. Biol. 2015, 889, 105–118. [Google Scholar] [CrossRef] [Green Version]
- Josson, S.; Gururajan, M.; Sung, S.Y.; Hu, P.; Shao, C.; Zhau, H.E.; Liu, C.; Lichterman, J.; Duan, P.; Li, Q.; et al. Stromal fibroblast-derived miR-409 promotes epithelial-to-mesenchymal transition and prostate tumorigenesis. Oncogene 2015, 34, 2690–2699. [Google Scholar] [CrossRef]
- Gururajan, M.; Josson, S.; Chu, G.C.; Lu, C.L.; Lu, Y.T.; Haga, C.L.; Zhau, H.E.; Liu, C.; Lichterman, J.; Duan, P.; et al. miR-154* and miR-379 in the DLK1-DIO3 microRNA mega-cluster regulate epithelial to mesenchymal transition and bone metastasis of prostate cancer. Clin. Cancer Res. 2014, 20, 6559–6569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruzzese, F.; Hägglöf, C.; Leone, A.; Sjöberg, E.; Roca, M.S.; Kiflemariam, S.; Sjöblom, T.; Hammarsten, P.; Egevad, L.; Bergh, A.; et al. Local and systemic protumorigenic effects of cancer-associated fibroblast-derived GDF15. Cancer Res. 2014, 74, 3408–3417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiao, S.L.; Chu, G.C.; Chung, L.W. Regulation of prostate cancer progression by the tumor microenvironment. Cancer Lett. 2016, 380, 340–348. [Google Scholar] [CrossRef] [Green Version]
- Loizzo, D.; Pandolfo, S.D.; Rogers, D.; Cerrato, C.; di Meo, N.A.; Autorino, R.; Mirone, V.; Ferro, M.; Porta, C.; Stella, A.; et al. Novel Insights into Autophagy and Prostate Cancer: A Comprehensive Review. Int. J. Mol. Sci. 2022, 23, 3826. [Google Scholar] [CrossRef]
- Zou, Z.; Tao, T.; Li, H.; Zhu, X. mTOR signaling pathway and mTOR inhibitors in cancer: Progress and challenges. Cell Biosci. 2020, 10, 31. [Google Scholar] [CrossRef] [Green Version]
- Giatromanolaki, A.; Sivridis, E.; Mendrinos, S.; Koutsopoulos, A.V.; Koukourakis, M.I. Autophagy proteins in prostate cancer: Relation with anaerobic metabolism and Gleason score. Urol. Oncol. 2014, 32, 39.e11–39.e18. [Google Scholar] [CrossRef] [PubMed]
- Aurilio, G.; Cimadamore, A.; Mazzucchelli, R.; Lopez-Beltran, A.; Verri, E.; Scarpelli, M.; Massari, F.; Cheng, L.; Santoni, M.; Montironi, R. Androgen Receptor Signaling Pathway in Prostate Cancer: From Genetics to Clinical Applications. Cells 2020, 9, 2653. [Google Scholar] [PubMed]
- Hu, J.; Chen, Q. The role of glucocorticoid receptor in prostate cancer progression: From bench to bedside. Int. Urol. Nephrol. 2017, 49, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef] [Green Version]
- Kurtz, D.M.; Soo, J.; Co Ting Keh, L.; Alig, S.; Chabon, J.J.; Sworder, B.J.; Schultz, A.; Jin, M.C.; Scherer, F.; Garofalo, A.; et al. Enhanced detection of minimal residual disease by targeted sequencing of phased variants in circulating tumor DNA. Nat. Biotechnol. 2021, 39, 1537–1547. [Google Scholar] [CrossRef]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Murtaza, M.; Dawson, S.-J.; Pogrebniak, K.; Rueda, O.M.; Provenzano, E.; Grant, J.; Chin, S.-F.; Tsui, D.W.Y.; Marass, F.; Gale, D.; et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat. Commun. 2015, 6, 8760. [Google Scholar] [CrossRef] [Green Version]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef]
- Campos-Fernández, E.; Barcelos, L.S.; de Souza, A.G.; Goulart, L.R.; Alonso-Goulart, V. Research landscape of liquid biopsies in prostate cancer. Am. J. Cancer Res. 2019, 9, 1309–1328. [Google Scholar]
- Wang, Y.; Wang, Z.; Gang, X.; Wang, G. Liquid biopsy in prostate cancer: Current status and future challenges of clinical application. Aging Male 2021, 24, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Wroclawski, M.L.; Serpa-Neto, A.; Fonseca, F.L.; Castro-Neves-Neto, O.; Pompeo, A.S.; Machado, M.T.; Pompeo, A.C.; del Giglio, A. Cell-free plasma DNA as biochemical biomarker for the diagnosis and follow-up of prostate cancer patients. Tumor Biol. 2013, 34, 2921–2927. [Google Scholar] [CrossRef] [PubMed]
- Bastian, P.J.; Palapattu, G.S.; Yegnasubramanian, S.; Lin, X.; Rogers, C.G.; Mangold, L.A.; Trock, B.; Eisenberger, M.; Partin, A.W.; Nelson, W.G. Prognostic Value of Preoperative Serum Cell-Free Circulating DNA in Men with Prostate Cancer Undergoing Radical Prostatectomy. Clin. Cancer Res. 2007, 13, 5361–5367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mouliere, F.; Smith, C.G.; Heider, K.; Su, J.; van der Pol, Y.; Thompson, M.; Morris, J.; Wan, J.C.M.; Chandrananda, D.; Hadfield, J.; et al. Fragmentation patterns and personalized sequencing of cell-free DNA in urine and plasma of glioma patients. EMBO Mol. Med. 2021, 13, e12881. [Google Scholar] [CrossRef]
- Van der Pol, Y.; Mouliere, F. Toward the Early Detection of Cancer by Decoding the Epigenetic and Environmental Fingerprints of Cell-Free DNA. Cancer Cell 2019, 36, 350–368. [Google Scholar] [CrossRef]
- Mouliere, F.; Chandrananda, D.; Piskorz, A.M.; Moore, E.K.; Morris, J.; Ahlborn, L.B.; Mair, R.; Goranova, T.; Marass, F.; Heider, K.; et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 2018, 10, eaat4921. [Google Scholar] [CrossRef] [Green Version]
- Underhill, H.R.; Kitzman, J.O.; Hellwig, S.; Welker, N.C.; Daza, R.; Baker, D.N.; Gligorich, K.M.; Rostomily, R.C.; Bronner, M.P.; Shendure, J. Fragment Length of Circulating Tumor DNA. PLoS Genet. 2016, 12, e1006162. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Gang, F.; Li, X.; Jin, T.; Houbao, H.; Yu, C.; Guorong, L. Plasma cell-free DNA and its DNA integrity as biomarker to distinguish prostate cancer from benign prostatic hyperplasia in patients with increased serum prostate-specific antigen. Int. Urol. Nephrol. 2013, 45, 1023–1028. [Google Scholar] [CrossRef]
- Khani, M.; Hosseini, J.; Mirfakhraie, R.; Habibi, M.; Azargashb, E.; Pouresmaeili, F. The value of the plasma circulating cell-free DNA concentration and integrity index as a clinical tool for prostate cancer diagnosis: A prospective case-control cohort study in an Iranian population. Cancer Manag. Res. 2019, 11, 4549–4556. [Google Scholar] [CrossRef] [Green Version]
- Ellinger, J.; Bastian, P.J.; Haan, K.I.; Heukamp, L.C.; Buettner, R.; Fimmers, R.; Mueller, S.C.; von Ruecker, A. Noncancerous PTGS2 DNA fragments of apoptotic origin in sera of prostate cancer patients qualify as diagnostic and prognostic indicators. Int. J. Cancer 2008, 122, 138–143. [Google Scholar] [CrossRef]
- Ponti, G.; Maccaferri, M.; Micali, S.; Manfredini, M.; Milandri, R.; Bianchi, G.; Pellacani, G.; Kaleci, S.; Chester, J.; Conti, A.; et al. Seminal Cell Free DNA Concentration Levels Discriminate between Prostate Cancer and Benign Prostatic Hyperplasia. Anticancer Res. 2018, 38, 5121–5125. [Google Scholar] [CrossRef] [PubMed]
- Romanel, A.; Gasi Tandefelt, D.; Conteduca, V.; Jayaram, A.; Casiraghi, N.; Wetterskog, D.; Salvi, S.; Amadori, D.; Zafeiriou, Z.; Rescigno, P.; et al. Plasma AR and abiraterone-resistant prostate cancer. Sci. Transl. Med. 2015, 7, 312re310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvi, S.; Casadio, V.; Conteduca, V.; Burgio, S.L.; Menna, C.; Bianchi, E.; Rossi, L.; Carretta, E.; Masini, C.; Amadori, D.; et al. Circulating cell-free AR and CYP17A1 copy number variations may associate with outcome of metastatic castration-resistant prostate cancer patients treated with abiraterone. Br. J. Cancer 2015, 112, 1717–1724. [Google Scholar] [CrossRef] [Green Version]
- Conteduca, V.; Wetterskog, D.; Sharabiani, M.T.A.; Grande, E.; Fernandez-Perez, M.P.; Jayaram, A.; Salvi, S.; Castellano, D.; Romanel, A.; Lolli, C.; et al. Androgen receptor gene status in plasma DNA associates with worse outcome on enzalutamide or abiraterone for castration-resistant prostate cancer: A multi-institution correlative biomarker study. Ann. Oncol. 2017, 28, 1508–1516. [Google Scholar] [CrossRef] [PubMed]
- Del Re, M.; Conteduca, V.; Crucitta, S.; Gurioli, G.; Casadei, C.; Restante, G.; Schepisi, G.; Lolli, C.; Cucchiara, F.; Danesi, R.; et al. Androgen receptor gain in circulating free DNA and splicing variant 7 in exosomes predict clinical outcome in CRPC patients treated with abiraterone and enzalutamide. Prostate Cancer Prostatic Dis. 2021, 24, 524–531. [Google Scholar] [CrossRef]
- Heitzer, E.; Ulz, P.; Belic, J.; Gutschi, S.; Quehenberger, F.; Fischereder, K.; Benezeder, T.; Auer, M.; Pischler, C.; Mannweiler, S.; et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013, 5, 30. [Google Scholar] [CrossRef] [Green Version]
- Kohli, M.; Tan, W.; Zheng, T.; Wang, A.; Montesinos, C.; Wong, C.; Du, P.; Jia, S.; Yadav, S.; Horvath, L.G.; et al. Clinical and genomic insights into circulating tumor DNA-based alterations across the spectrum of metastatic hormone-sensitive and castrate-resistant prostate cancer. EBioMedicine 2020, 54, 102728. [Google Scholar]
- Annala, M.; Vandekerkhove, G.; Khalaf, D.; Taavitsainen, S.; Beja, K.; Warner, E.W.; Sunderland, K.; Kollmannsberger, C.; Eigl, B.J.; Finch, D.; et al. Circulating Tumor DNA Genomics Correlate with Resistance to Abiraterone and Enzalutamide in Prostate Cancer. Cancer Discov. 2018, 8, 444–457. [Google Scholar] [CrossRef] [Green Version]
- Warner, E.; Herberts, C.; Fu, S.; Yip, S.; Wong, A.; Wang, G.; Ritch, E.; Murtha, A.J.; Vandekerkhove, G.; Fonseca, N.M.; et al. BRCA2, ATM, and CDK12 Defects Differentially Shape Prostate Tumor Driver Genomics and Clinical Aggression. Clin. Cancer Res. 2021, 27, 1650–1662. [Google Scholar] [CrossRef]
- Zhang, T.; Karsh, L.I.; Nissenblatt, M.J.; Canfield, S.E. Androgen Receptor Splice Variant, AR-V7, as a Biomarker of Resistance to Androgen Axis-Targeted Therapies in Advanced Prostate Cancer. Clin. Genitourin. Cancer 2020, 18, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Duffy, M.J. Biomarkers for prostate cancer: Prostate-specific antigen and beyond. Clin. Chem. Lab. Med. 2020, 58, 326–339. [Google Scholar] [CrossRef] [PubMed]
- Castro, E.; Jugurnauth-Little, S.; Karlsson, Q.; Al-Shahrour, F.; Piñeiro-Yañez, E.; Van de Poll, F.; Leongamornlert, D.; Dadaev, T.; Govindasami, K.; Guy, M.; et al. High burden of copy number alterations and c-MYC amplification in prostate cancer from BRCA2 germline mutation carriers. Ann. Oncol. 2015, 26, 2293–2300. [Google Scholar] [CrossRef] [PubMed]
- Seyedolmohadessin, S.M.; Akbari, M.T.; Nourmohammadi, Z.; Basiri, A.; Pourmand, G. Detection of Loss of Heterozygosity (LOH) Using Circulating Cell-free DNA (cfDNA) by Fluorescence-based Multiplex PCR for Identification of Patients With Prostate Cancer. Appl. Immunohistochem. Mol. Morphol. 2018, 26, 749–759. [Google Scholar] [CrossRef]
- Müller, I.; Urban, K.; Pantel, K.; Schwarzenbach, H. Comparison of Genetic Alterations Detected in Circulating Microsatellite DNA in Blood Plasma Samples of Patients with Prostate Cancer and Benign Prostatic Hyperplasia. Ann. N. Y. Acad. Sci. 2006, 1075, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Alix-Panabières, C.; Müller, I.; Letang, N.; Vendrell, J.-P.; Rebillard, X.; Pantel, K. Cell-free Tumor DNA in Blood Plasma As a Marker for Circulating Tumor Cells in Prostate Cancer. Clin. Cancer Res. 2009, 15, 1032–1038. [Google Scholar] [CrossRef] [Green Version]
- Lau, E.; McCoy, P.; Reeves, F.; Chow, K.; Clarkson, M.; Kwan, E.M.; Packwood, K.; Northen, H.; He, M.; Kingsbury, Z.; et al. Detection of ctDNA in plasma of patients with clinically localised prostate cancer is associated with rapid disease progression. Genome Med. 2020, 12, 72. [Google Scholar] [CrossRef]
- Hennigan, S.T.; Trostel, S.Y.; Terrigino, N.T.; Voznesensky, O.S.; Schaefer, R.J.; Whitlock, N.C.; Wilkinson, S.; Carrabba, N.V.; Atway, R.; Shema, S.; et al. Low Abundance of Circulating Tumor DNA in Localized Prostate Cancer. JCO Precis. Oncol. 2019, 3. [Google Scholar] [CrossRef]
- Baden, J.; Adams, S.; Astacio, T.; Jones, J.; Markiewicz, J.; Painter, J.; Trust, C.; Wang, Y.; Green, G. Predicting prostate biopsy result in men with prostate specific antigen 2.0 to 10.0 ng/ml using an investigational prostate cancer methylation assay. J. Urol. 2011, 186, 2101–2106. [Google Scholar] [CrossRef]
- Bastian, P.J.; Palapattu, G.S.; Lin, X.; Yegnasubramanian, S.; Mangold, L.A.; Trock, B.; Eisenberger, M.A.; Partin, A.W.; Nelson, W.G. Preoperative serum DNA GSTP1 CpG island hypermethylation and the risk of early prostate-specific antigen recurrence following radical prostatectomy. Clin. Cancer Res. 2005, 11, 4037–4043. [Google Scholar] [CrossRef] [Green Version]
- Cairns, P.; Esteller, M.; Herman, J.G.; Schoenberg, M.; Jeronimo, C.; Sanchez-Cespedes, M.; Chow, N.H.; Grasso, M.; Wu, L.; Westra, W.B.; et al. Molecular detection of prostate cancer in urine by GSTP1 hypermethylation. Clin. Cancer Res. 2001, 7, 2727–2730. [Google Scholar]
- Hendriks, R.J.; Dijkstra, S.; Smit, F.P.; Vandersmissen, J.; Van de Voorde, H.; Mulders, P.F.A.; van Oort, I.M.; Van Criekinge, W.; Schalken, J.A. Epigenetic markers in circulating cell-free DNA as prognostic markers for survival of castration-resistant prostate cancer patients. Prostate 2018, 78, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Mahon, K.L.; Qu, W.; Devaney, J.; Paul, C.; Castillo, L.; Wykes, R.J.; Chatfield, M.D.; Boyer, M.J.; Stockler, M.R.; Marx, G.; et al. Methylated Glutathione S-transferase 1 (mGSTP1) is a potential plasma free DNA epigenetic marker of prognosis and response to chemotherapy in castrate-resistant prostate cancer. Br. J. Cancer 2014, 111, 1802–1809. [Google Scholar] [CrossRef] [PubMed]
- Moreira-Barbosa, C.; Barros-Silva, D.; Costa-Pinheiro, P.; Torres-Ferreira, J.; Constâncio, V.; Freitas, R.; Oliveira, J.; Antunes, L.; Henrique, R.; Jerónimo, C. Comparing diagnostic and prognostic performance of two-gene promoter methylation panels in tissue biopsies and urines of prostate cancer patients. Clin. Epigenetics 2018, 10, 132. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, E.; Tuzova, A.V.; Walsh, A.L.; Russell, N.M.; O’Brien, O.; Kelly, S.; Dhomhnallain, O.N.; DeBarra, L.; Dale, C.M.; Brugman, R.; et al. epiCaPture: A Urine DNA Methylation Test for Early Detection of Aggressive Prostate Cancer. JCO Precis Oncol. 2019, 2019. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Olkhov-Mitsel, E.; van der Kwast, T.; Sykes, J.; Zdravic, D.; Venkateswaran, V.; Zlotta, A.R.; Loblaw, A.; Fleshner, N.E.; Klotz, L.; et al. Urinary DNA Methylation Biomarkers for Noninvasive Prediction of Aggressive Disease in Patients with Prostate Cancer on Active Surveillance. J. Urol. 2017, 197, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Gordevičius, J.; Kriščiūnas, A.; Groot, D.E.; Yip, S.M.; Susic, M.; Kwan, A.; Kustra, R.; Joshua, A.M.; Chi, K.N.; Petronis, A.; et al. Cell-Free DNA Modification Dynamics in Abiraterone Acetate-Treated Prostate Cancer Patients. Clin. Cancer Res. 2018, 24, 3317–3324. [Google Scholar] [CrossRef] [Green Version]
- Moss, J.; Magenheim, J.; Neiman, D.; Zemmour, H.; Loyfer, N.; Korach, A.; Samet, Y.; Maoz, M.; Druid, H.; Arner, P.; et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 2018, 9, 5068. [Google Scholar] [CrossRef] [Green Version]
- Silva, R.; Moran, B.; Russell, N.M.; Fahey, C.; Vlajnic, T.; Manecksha, R.P.; Finn, S.P.; Brennan, D.J.; Gallagher, W.M.; Perry, A.S. Evaluating liquid biopsies for methylomic profiling of prostate cancer. Epigenetics 2020, 15, 715–727. [Google Scholar] [CrossRef] [Green Version]
- Gerhauser, C.; Favero, F.; Risch, T.; Simon, R.; Feuerbach, L.; Assenov, Y.; Heckmann, D.; Sidiropoulos, N.; Waszak, S.M.; Hübschmann, D.; et al. Molecular Evolution of Early-Onset Prostate Cancer Identifies Molecular Risk Markers and Clinical Trajectories. Cancer Cell 2018, 34, 996–1011.e1018. [Google Scholar] [CrossRef] [Green Version]
- Shen, S.Y.; Burgener, J.M.; Bratman, S.V.; De Carvalho, D.D. Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA. Nat. Protoc. 2019, 14, 2749–2780. [Google Scholar] [CrossRef]
- Shen, S.Y.; Singhania, R.; Fehringer, G.; Chakravarthy, A.; Roehrl, M.H.A.; Chadwick, D.; Zuzarte, P.C.; Borgida, A.; Wang, T.T.; Li, T.; et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018, 563, 579–583. [Google Scholar] [CrossRef] [PubMed]
- Chemi, F.; Pearce, S.P.; Clipson, A.; Hill, S.M.; Conway, A.M.; Richardson, S.A.; Kamieniecka, K.; Caeser, R.; White, D.J.; Mohan, S.; et al. cfDNA methylome profiling for detection and subtyping of small cell lung cancers. Nat Cancer 2022, 3, 1260–1270. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Soupir, A.C.; Wang, L. Cell-free DNA methylome profiling by MBD-seq with ultra-low input. Epigenetics 2022, 17, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Siejka-Zielińska, P.; Cheng, J.; Jackson, F.; Liu, Y.; Soonawalla, Z.; Reddy, S.; Silva, M.; Puta, L.; McCain, M.V.; Culver, E.L.; et al. Cell-free DNA TAPS provides multimodal information for early cancer detection. Sci. Adv. 2021, 7, eabh0534. [Google Scholar] [CrossRef]
- Stackpole, M.L.; Zeng, W.; Li, S.; Liu, C.-C.; Zhou, Y.; He, S.; Yeh, A.; Wang, Z.; Sun, F.; Li, Q.; et al. Cost-effective methylome sequencing of cell-free DNA for accurately detecting and locating cancer. Nat. Commun. 2022, 13, 5566. [Google Scholar] [CrossRef]
- Chen, S.; Petricca, J.; Ye, W.; Guan, J.; Zeng, Y.; Cheng, N.; Gong, L.; Shen, S.Y.; Hua, J.T.; Crumbaker, M.; et al. The cell-free DNA methylome captures distinctions between localized and metastatic prostate tumors. Nat. Commun. 2022, 13, 6467. [Google Scholar] [CrossRef] [PubMed]
- Roest, H.P.; JNM, I.J.; van der Laan, L.J.W. Evaluation of RNA isolation methods for microRNA quantification in a range of clinical biofluids. BMC Biotechnol. 2021, 21, 48. [Google Scholar] [CrossRef]
- McKiernan, J.; Donovan, M.J.; O’Neill, V.; Bentink, S.; Noerholm, M.; Belzer, S.; Skog, J.; Kattan, M.W.; Partin, A.; Andriole, G.; et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol. 2016, 2, 882–889. [Google Scholar] [CrossRef] [Green Version]
- Tomlins, S.A.; Day, J.R.; Lonigro, R.J.; Hovelson, D.H.; Siddiqui, J.; Kunju, L.P.; Dunn, R.L.; Meyer, S.; Hodge, P.; Groskopf, J.; et al. Urine TMPRSS2:ERG Plus PCA3 for Individualized Prostate Cancer Risk Assessment. Eur. Urol. 2016, 70, 45–53. [Google Scholar] [CrossRef] [Green Version]
- Van Neste, L.; Hendriks, R.J.; Dijkstra, S.; Trooskens, G.; Cornel, E.B.; Jannink, S.A.; de Jong, H.; Hessels, D.; Smit, F.P.; Melchers, W.J.; et al. Detection of High-grade Prostate Cancer Using a Urinary Molecular Biomarker-Based Risk Score. Eur. Urol. 2016, 70, 740–748. [Google Scholar] [CrossRef]
- Souza, M.F.; Kuasne, H.; Barros-Filho, M.C.; Cilião, H.L.; Marchi, F.A.; Fuganti, P.E.; Rogatto, S.R.; Cólus, I.M.S. Circulating mRNA signature as a marker for high-risk prostate cancer. Carcinogenesis 2020, 41, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Mihelich, B.L.; Maranville, J.C.; Nolley, R.; Peehl, D.M.; Nonn, L. Elevated serum microRNA levels associate with absence of high-grade prostate cancer in a retrospective cohort. PLoS ONE 2015, 10, e0124245. [Google Scholar] [CrossRef]
- Fredsøe, J.; Rasmussen, A.K.I.; Thomsen, A.R.; Mouritzen, P.; Høyer, S.; Borre, M.; Ørntoft, T.F.; Sørensen, K.D. Diagnostic and Prognostic MicroRNA Biomarkers for Prostate Cancer in Cell-free Urine. Eur. Urol. Focus 2018, 4, 825–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooperberg, M.R.; Pasta, D.J.; Elkin, E.P.; Litwin, M.S.; Latini, D.M.; Du Chane, J.; Carroll, P.R. The University of California, San Francisco Cancer of the Prostate Risk Assessment score: A straightforward and reliable preoperative predictor of disease recurrence after radical prostatectomy. J. Urol. 2005, 173, 1938–1942. [Google Scholar] [CrossRef] [Green Version]
- Shen, J.; Hruby, G.W.; McKiernan, J.M.; Gurvich, I.; Lipsky, M.J.; Benson, M.C.; Santella, R.M. Dysregulation of circulating microRNAs and prediction of aggressive prostate cancer. Prostate 2012, 72, 1469–1477. [Google Scholar] [CrossRef] [Green Version]
- Hoey, C.; Ahmed, M.; Fotouhi Ghiam, A.; Vesprini, D.; Huang, X.; Commisso, K.; Commisso, A.; Ray, J.; Fokas, E.; Loblaw, D.A.; et al. Circulating miRNAs as non-invasive biomarkers to predict aggressive prostate cancer after radical prostatectomy. J. Transl. Med. 2019, 17, 173. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.T.; Shi, T.; Srivastava, S.; Kagan, J.; Liu, T.; Rodland, K.D. Proteomic Analysis of Exosomes for Discovery of Protein Biomarkers for Prostate and Bladder Cancer. Cancers 2020, 12, 2335. [Google Scholar] [CrossRef]
- Nilsson, J.; Skog, J.; Nordstrand, A.; Baranov, V.; Mincheva-Nilsson, L.; Breakefield, X.O.; Widmark, A. Prostate cancer-derived urine exosomes: A novel approach to biomarkers for prostate cancer. Br. J. Cancer 2009, 100, 1603–1607. [Google Scholar] [CrossRef]
- Zabegina, L.; Nazarova, I.; Nikiforova, N.; Slyusarenko, M.; Sidina, E.; Knyazeva, M.; Tsyrlina, E.; Novikov, S.; Reva, S.; Malek, A. A New Approach for Prostate Cancer Diagnosis by miRNA Profiling of Prostate-Derived Plasma Small Extracellular Vesicles. Cells 2021, 10, 2372. [Google Scholar]
- Woo, J.; Santasusagna, S.; Banks, J.; Pastor-Lopez, S.; Yadav, K.; Carceles-Cordon, M.; Dominguez-Andres, A.; Den, R.B.; Languino, L.R.; Pippa, R.; et al. Urine Extracellular Vesicle GATA2 mRNA Discriminates Biopsy Result in Men with Suspicion of Prostate Cancer. J. Urol. 2020, 204, 691–700. [Google Scholar] [CrossRef]
- Dhondt, B.; Geeurickx, E.; Tulkens, J.; Van Deun, J.; Vergauwen, G.; Lippens, L.; Miinalainen, I.; Rappu, P.; Heino, J.; Ost, P.; et al. Unravelling the proteomic landscape of extracellular vesicles in prostate cancer by density-based fractionation of urine. J. Extracell. Vesicles 2020, 9, 1736935. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Nonomura, N. Urinary biomarkers of prostate cancer. Int. J. Urol. 2018, 25, 770–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasseur, A.; Kiavue, N.; Bidard, F.C.; Pierga, J.Y.; Cabel, L. Clinical utility of circulating tumor cells: An update. Mol. Oncol. 2021, 15, 1647–1666. [Google Scholar] [CrossRef] [PubMed]
- Boya, M.; Chu, C.H.; Liu, R.; Ozkaya-Ahmadov, T.; Sarioglu, A.F. Circulating Tumor Cell Enrichment Technologies. In Tumor Liquid Biopsies; Recent Results in Cancer Research Book Series; Springer: Berlin/Heidelberg, Germany, 2020; Volume 215, pp. 25–55. [Google Scholar] [CrossRef]
- Malihi, P.D.; Graf, R.P.; Rodriguez, A.; Ramesh, N.; Lee, J.; Sutton, R.; Jiles, R.; Ruiz Velasco, C.; Sei, E.; Kolatkar, A.; et al. Single-Cell Circulating Tumor Cell Analysis Reveals Genomic Instability as a Distinctive Feature of Aggressive Prostate Cancer. Clin. Cancer Res. 2020, 26, 4143–4153. [Google Scholar] [CrossRef]
- Wang, X.; Grasso, C.S.; Jordahl, K.M.; Kolb, S.; Nyame, Y.A.; Wright, J.L.; Ostrander, E.A.; Troyer, D.A.; Lance, R.; Feng, Z.; et al. Copy number alterations are associated with metastatic-lethal progression in prostate cancer. Prostate Cancer Prostatic Dis. 2020, 23, 494–506. [Google Scholar] [CrossRef]
- Harshman, L.C.; Wang, V.X.; Hamid, A.A.; Santone, G.; Drake, C.G.; Carducci, M.A.; DiPaola, R.S.; Fichorova, R.N.; Sweeney, C.J. Impact of baseline serum IL-8 on metastatic hormone-sensitive prostate cancer outcomes in the Phase 3 CHAARTED trial (E3805). Prostate 2020, 80, 1429–1437. [Google Scholar] [CrossRef]
- Gong, Y.; Chippada-Venkata, U.D.; Oh, W.K. Roles of matrix metalloproteinases and their natural inhibitors in prostate cancer progression. Cancers 2014, 6, 1298–1327. [Google Scholar] [CrossRef] [Green Version]
- Shajarehpoor Salavati, L.; Tafvizi, F.; Manjili, H.K. The association between MMP2 -1306 C > T (rs243865) polymorphism and risk of prostate cancer. Ir. J. Med. Sci. 2017, 186, 103–111. [Google Scholar] [CrossRef]
- Dong, M.; Lih, T.M.; Chen, S.Y.; Cho, K.C.; Eguez, R.V.; Höti, N.; Zhou, Y.; Yang, W.; Mangold, L.; Chan, D.W.; et al. Urinary glycoproteins associated with aggressive prostate cancer. Theranostics 2020, 10, 11892–11907. [Google Scholar] [CrossRef]
- Di Minno, A.; Aveta, A.; Gelzo, M.; Tripodi, L.; Pandolfo, S.D.; Crocetto, F.; Imbimbo, C.; Castaldo, G. 8-Hydroxy-2-Deoxyguanosine and 8-Iso-Prostaglandin F2α: Putative Biomarkers to assess Oxidative Stress Damage Following Robot-Assisted Radical Prostatectomy (RARP). J. Clin. Med. 2022, 11, 6102. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Liquid biopsy and minimal residual disease—Latest advances and implications for cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, Z.; Wang, Q.; Chang, K.; Zhang, J.; Ye, D.; Kong, Y.; Dai, B. Presence of CD133-positive circulating tumor cells predicts worse progression-free survival in patients with metastatic castration-sensitive prostate cancer. Int. J. Urol. 2022, 29, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Broncy, L.; Paterlini-Bréchot, P. Clinical Impact of Circulating Tumor Cells in Patients with Localized Prostate Cancer. Cells 2019, 8, 676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Martinez, A.; Simon-Saez, I.; Perales, S.; Garrido-Navas, C.; Russo, A.; de Miguel-Perez, D.; Puche-Sanz, I.; Alaminos, C.; Ceron, J.; Lorente, J.A.; et al. Exchange of cellular components between platelets and tumor cells: Impact on tumor cells behavior. Theranostics 2022, 12, 2150–2161. [Google Scholar] [CrossRef]
- Clar, K.L.; Hinterleitner, C.; Schneider, P.; Salih, H.R.; Maurer, S. Inhibition of NK Reactivity against Solid Tumors by Platelet-Derived RANKL. Cancers 2019, 11, 277. [Google Scholar] [CrossRef] [Green Version]
- Tjon-Kon-Fat, L.A.; Lundholm, M.; Schröder, M.; Wurdinger, T.; Thellenberg-Karlsson, C.; Widmark, A.; Wikström, P.; Nilsson, R.J.A. Platelets harbor prostate cancer biomarkers and the ability to predict therapeutic response to abiraterone in castration resistant patients. Prostate 2018, 78, 48–53. [Google Scholar] [CrossRef]
- TracerX. TRAcking Cancer Evolution through Therapy (Rx). Available online: http://tracerx.co.uk/studies/prostate/ (accessed on 26 November 2022).
- Turajlic, S.; Sottoriva, A.; Graham, T.; Swanton, C. Resolving genetic heterogeneity in cancer. Nat. Rev. Genet 2019, 20, 404–416. [Google Scholar] [CrossRef]
- Xi, J.; Yuan, X.; Wang, M.; Li, A.; Li, X.; Huang, Q. Inferring subgroup-specific driver genes from heterogeneous cancer samples via subspace learning with subgroup indication. Bioinformatics 2020, 36, 1855–1863. [Google Scholar] [CrossRef]
- Liu, S.; Wang, W.; Zhao, Y.; Liang, K.; Huang, Y. Identification of Potential Key Genes for Pathogenesis and Prognosis in Prostate Cancer by Integrated Analysis of Gene Expression Profiles and the Cancer Genome Atlas. Front. Oncol. 2020, 10, 809. [Google Scholar] [CrossRef]
- The Cancer Gene Census (CGC). Available online: https://cancer.sanger.ac.uk/census (accessed on 26 November 2022).
- Kesch, C.; Radtke, J.P.; Wintsche, A.; Wiesenfarth, M.; Luttje, M.; Gasch, C.; Dieffenbacher, S.; Pecqueux, C.; Teber, D.; Hatiboglu, G.; et al. Correlation between genomic index lesions and mpMRI and (68)Ga-PSMA-PET/CT imaging features in primary prostate cancer. Sci. Rep. 2018, 8, 16708. [Google Scholar] [CrossRef] [Green Version]
- Norris, J.M.; Simpson, B.S.; Parry, M.A.; Allen, C.; Ball, R.; Freeman, A.; Kelly, D.; Kirkham, A.; Kasivisvanathan, V.; Whitaker, H.C.; et al. Genetic landscape of prostate cancer conspicuity on multiparametric MRI: A protocol for a systematic review and bioinformatic analysis. BMJ Open 2020, 10, e034611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrison, G.; Buckley, J.; Ostrow, D.; Varghese, B.; Cen, S.Y.; Werbin, J.; Ericson, N.; Cunha, A.; Lu, Y.T.; George, T.; et al. Non-Invasive Profiling of Advanced Prostate Cancer via Multi-Parametric Liquid Biopsy and Radiomic Analysis. Int. J. Mol. Sci. 2022, 23, 2571. [Google Scholar] [CrossRef] [PubMed]
- Alarcón-Zendejas, A.P.; Scavuzzo, A.; Jiménez-Ríos, M.A.; Álvarez-Gómez, R.M.; Montiel-Manríquez, R.; Castro-Hernández, C.; Jiménez-Dávila, M.A.; Pérez-Montiel, D.; González-Barrios, R.; Jiménez-Trejo, F.; et al. The promising role of new molecular biomarkers in prostate cancer: From coding and non-coding genes to artificial intelligence approaches. Prostate Cancer Prostatic Dis. 2022, 25, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Schwartzberg, L.S.; Li, G.; Tolba, K.; Bourla, A.B.; Schulze, K.; Gadgil, R.; Fine, A.; Lofgren, K.T.; Graf, R.P.; Oxnard, G.R.; et al. Complementary Roles for Tissue- and Blood-Based Comprehensive Genomic Profiling for Detection of Actionable Driver Alterations in Advanced NSCLC. JTO Clin. Res. Rep. 2022, 3, 100386. [Google Scholar] [CrossRef] [PubMed]
- Peneder, P.; Stutz, A.M.; Surdez, D.; Krumbholz, M.; Semper, S.; Chicard, M.; Sheffield, N.C.; Pierron, G.; Lapouble, E.; Totzl, M.; et al. Multimodal analysis of cell-free DNA whole-genome sequencing for pediatric cancers with low mutational burden. Nat. Commun. 2021, 12, 3230. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Park, Y.; Kim, S. Towards multi-omics characterization of tumor heterogeneity: A comprehensive review of statistical and machine learning approaches. Brief. Bioinform. 2021, 22, bbaa188. [Google Scholar] [CrossRef]
- Onuchic, V.; Hartmaier, R.J.; Boone, D.N.; Samuels, M.L.; Patel, R.Y.; White, W.M.; Garovic, V.D.; Oesterreich, S.; Roth, M.E.; Lee, A.V.; et al. Epigenomic Deconvolution of Breast Tumors Reveals Metabolic Coupling between Constituent Cell Types. Cell Rep. 2016, 17, 2075–2086. [Google Scholar] [CrossRef] [Green Version]
- De Laere, B.; Oeyen, S.; Mayrhofer, M.; Whitington, T.; van Dam, P.J.; Van Oyen, P.; Ghysel, C.; Ampe, J.; Ost, P.; Demey, W.; et al. TP53 Outperforms Other Androgen Receptor Biomarkers to Predict Abiraterone or Enzalutamide Outcome in Metastatic Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2019, 25, 1766–1773. [Google Scholar] [CrossRef] [Green Version]
- De Laere, B.; van Dam, P.J.; Whitington, T.; Mayrhofer, M.; Diaz, E.H.; Van den Eynden, G.; Vandebroek, J.; Del-Favero, J.; Van Laere, S.; Dirix, L.; et al. Comprehensive Profiling of the Androgen Receptor in Liquid Biopsies from Castration-resistant Prostate Cancer Reveals Novel Intra-AR Structural Variation and Splice Variant Expression Patterns. Eur. Urol. 2017, 72, 192–200. [Google Scholar] [CrossRef]
- Kohli, M.; Li, J.; Du, M.; Hillman, D.W.; Dehm, S.M.; Tan, W.; Carlson, R.; Campion, M.B.; Wang, L.; Wang, L.; et al. Prognostic association of plasma cell-free DNA-based androgen receptor amplification and circulating tumor cells in pre-chemotherapy metastatic castration-resistant prostate cancer patients. Prostate Cancer Prostatic Dis. 2018, 21, 411–418. [Google Scholar] [CrossRef]
- Hofmann, L.; Sallinger, K.; Haudum, C.; Smolle, M.; Heitzer, E.; Moser, T.; Novy, M.; Gesson, K.; Kroneis, T.; Bauernhofer, T.; et al. A Multi-Analyte Approach for Improved Sensitivity of Liquid Biopsies in Prostate Cancer. Cancers 2020, 12, 2247. [Google Scholar] [CrossRef] [PubMed]
- Fettke, H.; Kwan, E.M.; Docanto, M.M.; Bukczynska, P.; Ng, N.; Graham, L.K.; Mahon, K.; Hauser, C.; Tan, W.; Wang, X.H.; et al. Combined Cell-free DNA and RNA Profiling of the Androgen Receptor: Clinical Utility of a Novel Multianalyte Liquid Biopsy Assay for Metastatic Prostate Cancer. Eur. Urol. 2020, 78, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Connell, S.P.; O’Reilly, E.; Tuzova, A.; Webb, M.; Hurst, R.; Mills, R.; Zhao, F.; Bapat, B.; Cooper, C.S.; Perry, A.S.; et al. Development of a multivariable risk model integrating urinary cell DNA methylation and cell-free RNA data for the detection of significant prostate cancer. Prostate 2020, 80, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Burgener, J.M.; Zou, J.; Zhao, Z.; Zheng, Y.; Shen, S.Y.; Huang, S.H.; Keshavarzi, S.; Xu, W.; Liu, F.F.; Liu, G.; et al. Tumor-Naive Multimodal Profiling of Circulating Tumor DNA in Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res. 2021, 27, 4230–4244. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eickelschulte, S.; Riediger, A.L.; Angeles, A.K.; Janke, F.; Duensing, S.; Sültmann, H.; Görtz, M. Biomarkers for the Detection and Risk Stratification of Aggressive Prostate Cancer. Cancers 2022, 14, 6094. https://doi.org/10.3390/cancers14246094
Eickelschulte S, Riediger AL, Angeles AK, Janke F, Duensing S, Sültmann H, Görtz M. Biomarkers for the Detection and Risk Stratification of Aggressive Prostate Cancer. Cancers. 2022; 14(24):6094. https://doi.org/10.3390/cancers14246094
Chicago/Turabian StyleEickelschulte, Samaneh, Anja Lisa Riediger, Arlou Kristina Angeles, Florian Janke, Stefan Duensing, Holger Sültmann, and Magdalena Görtz. 2022. "Biomarkers for the Detection and Risk Stratification of Aggressive Prostate Cancer" Cancers 14, no. 24: 6094. https://doi.org/10.3390/cancers14246094