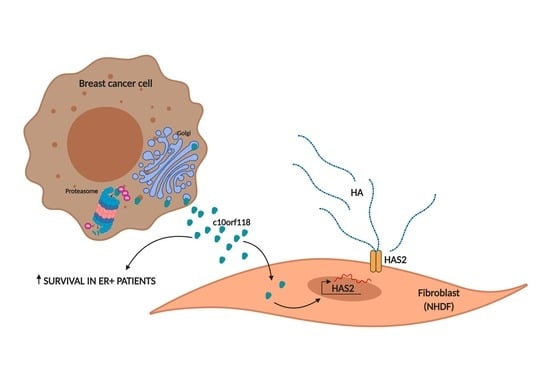
The Secreted Protein C10orf118 Is a New Regulator of Hyaluronan Synthesis Involved in Tumour-Stroma Cross-Talk
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. A Soluble Factor in 8701-BC Cells Conditioned Medium Induces Expression of HAS2 in NHDF
2.2. C10orf118 Is Found Both in Cell Lysate and Secreted Media of Different Breast Cancer Cell Lines
2.3. Endogenous c10orf118 Localizes in the Golgi Apparatus in MCF-7 Cells
2.4. Effects of C10orf118 Silencing in MCF-7 Cells
2.5. c10orf118 Silencing Influences HA-Related Genes Expression in MCF-7
2.6. Specificity of c10orf118 Protein Action in Inducing HA Synthesis by Stromal Cells
2.7. C10orf118 Is Detected in Breast Cancer Cell Tissues and Is Estimated to Associate with Estrogen Receptor (ER) Expression
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Tissue Specimens
4.2. Isolation of a Soluble Factor from 8701-BC Cell Conditioned Medium by MALDI/MS
4.3. NHDF Treatment with 8701-BC Conditioned Medium
4.4. SDS-PAGE and Western Blot
4.5. Immunoprecipitation
4.6. Immunofluorescence and Confocal Microscopy
4.7. Cell Viability Assay (MTT)
4.8. Wound Healing Assay
4.9. C10orf118 Overexpression
4.10. RNA Extraction and cDNA Synthesis
4.11. Quantitative RT-PCR
4.12. Transwell System
4.13. HPLC Analysis for HA Disaccharides Determination in Culture Medium
4.14. Neutralization of c10orf118 by a Specific Antibody in Conditioned Medium of MCF-7 Cells
4.15. Cell Treatment with c10orf118 Recombinant Protein
4.16. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barcellos-Hoff, M.H.; Lyden, D.; Wang, T.C. The evolution of the cancer niche during multistage carcinogenesis. Nat. Rev. Cancer 2013, 13, 511–518. [Google Scholar] [CrossRef]
- Xing, Y.; Zhao, S.; Zhou, B.P.; Mi, J. Metabolic reprogramming of the tumour microenvironment. FEBS J. 2015, 282, 3892–3898. [Google Scholar] [CrossRef] [PubMed]
- Lasorella, A.; Benezra, R.; Iavarone, A. The ID proteins: Master regulators of cancer stem cells and tumour aggressiveness. Nat. Rev. Cancer 2014, 14, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Saidou, J.; Watabe, K. Cancer associated fibroblasts (CAFs) in tumor microenvironment. Front. Biosci. 2010, 15, 166–179. [Google Scholar] [CrossRef] [Green Version]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sreekumar, A.; Poisson, L.M.; Rajendiran, T.M.; Khan, A.P.; Cao, Q.; Yu, J.; Laxman, B.; Mehra, R.; Lonigro, R.J.; Li, Y.; et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009, 457, 910–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Huang, P. Cancer-stromal interactions role in cell survival, metabolism and drug sensitivity. Cancer Biol. Ther. 2011, 11, 150–156. [Google Scholar] [CrossRef] [Green Version]
- Louault, K.; Bonneaud, T.L.; Séveno, C.; Gomez-Bougie, P.; Nguyen, F.; Gautier, F.; Bourgeois, N.; Loussouarn, D.; Kerdraon, O.; Barillé-Nion, S.; et al. Interactions between cancer-associated fibroblasts and tumor cells promote MCL-1 dependency in estrogen receptor-positive breast cancers. Oncogene 2019, 38, 3261–3273. [Google Scholar] [CrossRef] [Green Version]
- Caon, I.; Bartolini, B.; Parnigoni, A.; Caravà, E.; Moretto, P.; Viola, M.; Karousou, E.; Vigetti, D.; Passi, A. Revisiting the hallmarks of cancer: The role of hyaluronan. Semin. Cancer Biol. 2019, 62, 9–19. [Google Scholar] [CrossRef]
- Filpa, V.; Bistoletti, M.; Caon, I.; Moro, E.; Grimaldi, A.; Moretto, P.; Baj, A.; Giron, M.C.; Karousou, E.; Viola, M.; et al. Changes in hyaluronan deposition in the rat myenteric plexus after experimentally-induced colitis. Sci. Rep. 2017, 7, 17644. [Google Scholar] [CrossRef]
- Karousou, E.; Misra, S.; Ghatak, S.; Dobra, K.; Götte, M.; Vigetti, D.; Passi, A.; Karamanos, N.K.; Skandalis, S.S. Roles and targeting of the HAS/hyaluronan/CD44 molecular system in cancer. Matrix Biol. 2017, 59, 3–22. [Google Scholar] [CrossRef]
- Tolg, C.; McCarthy, J.B.; Yazdani, A.; Turley, E.A. Hyaluronan and RHAMM in wound repair and the "cancerization" of stromal tissues. Biomed. Res. Int. 2014, 2014, 103923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourguignon, L.Y.W.; Wong, G.; Earle, C.; Krueger, K.; Spevak, C.C. Hyaluronan-CD44 interaction promotes c-Src-mediated twist signaling, microRNA-10b expression, and RhoA/RhoC up-regulation, leading to Rho-kinase-associated cytoskeleton activation and breast tumor cell invasion. J. Biol. Chem. 2010, 285, 36721–36735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toole, B.P. Hyaluronan: From extracellular glue to pericellular cue. Nat. Rev. Cancer 2004, 4, 528–539. [Google Scholar] [CrossRef]
- Tavianatou, A.G.; Caon, I.; Franchi, M.; Piperigkou, Z.; Galesso, D.; Karamanos, N.K. Hyaluronan: Molecular size-dependent signaling and biological functions in inflammation and cancer. FEBS J. 2019, 2883–2908. [Google Scholar] [CrossRef] [PubMed]
- Heldin, P.; Basu, K.; Olofsson, B.; Porsch, H.; Kozlova, I.; Kahata, K. Deregulation of hyaluronan synthesis, degradation and binding promotes breast cancer. J. Biochem. 2013, 154, 395–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heldin, P. Differential synthesis and binding of hyaluronan by human breast cancer cell lines: Relationship to hormone receptor status. Oncol. Rep. 1996, 3, 1011–1016. [Google Scholar] [CrossRef]
- Li, Y.; Li, L.; Brown, T.J.; Heldin, P. Silencing of hyaluronan synthase 2 suppresses the malignant phenotype of invasive breast cancer cells. Int. J. Cancer 2007, 120, 2557–2567. [Google Scholar] [CrossRef]
- Passi, A.; Vigetti, D.; Buraschi, S.; Iozzo, R.V. Dissecting the role of hyaluronan synthases in the tumor microenvironment. FEBS J. 2019, febs.14847. [Google Scholar] [CrossRef] [PubMed]
- Auvinen, P.; Tammi, R.; Parkkinen, J.; Tammi, M.; ÅGren, U.; Johansson, R.; Hirvikoski, P.; Eskelinen, M.; Kosma, V.-M. Hyaluronan in Peritumoral Stroma and Malignant Cells Associates with Breast Cancer Spreading and Predicts Survival. Am. J. Pathol. 2000, 156, 529–536. [Google Scholar] [CrossRef]
- Porsch, H.; Mehić, M.; Olofsson, B.; Heldin, P.; Heldin, C.H. Platelet-derived growth factor β-receptor, transforming growth factor β type I receptor, and CD44 protein modulate each other’s signaling and stability. J. Biol. Chem. 2014, 289, 19747–19757. [Google Scholar] [CrossRef] [Green Version]
- Chanmee, T.; Ontong, P.; Izumikawa, T.; Higashide, M.; Mochizuki, N.; Chokchaitaweesuk, C.; Khansai, M.; Nakajima, K.; Kakizaki, I.; Kongtawelert, P.; et al. Hyaluronan production regulates metabolic and cancer stem-like properties of breast cancer cells via hexosamine biosynthetic pathway-coupled HIF-1 signaling. J. Biol. Chem. 2016, 291, 24105–24120. [Google Scholar] [CrossRef] [Green Version]
- Caon, I.; Parnigoni, A.; Viola, M.; Karousou, E.; Passi, A.; Vigetti, D. Cell Energy Metabolism and Hyaluronan Synthesis. J. Histochem. Cytochem. 2021, 69, 35–47. [Google Scholar] [CrossRef]
- Huet, E.; Jaroz, C.; Nguyen, H.Q.; Belkacemi, Y.; Taille, A.; Stavrinides, V.; Whitaker, H. Stroma in normal and cancer wound healing. FEBS J. 2019, 286, 2909–2920. [Google Scholar] [CrossRef] [Green Version]
- Gillingham, A.K.; Sinka, R.; Torres, I.L.; Lilley, K.S.; Munro, S. Toward a Comprehensive Map of the Effectors of Rab GTPases. Dev. Cell 2014, 31, 358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muschalik, N.; Munro, S. Golgins. Curr. Biol. 2018, 28, R374–R376. [Google Scholar] [CrossRef] [Green Version]
- Cattin-Ortolá, J.; Topalidou, I.; Lau, H.T.; Hummer, B.; Asensio, C.S.; Ong, S.E.; Ailion, M. CCDC186 controls dense-core vesicle cargo sorting by exit. bioRxiv 2019, 616458. [Google Scholar] [CrossRef]
- Renoir, J.M.; Marsaud, V.; Lazennec, G. Estrogen receptor signaling as a target for novel breast cancer therapeutics. Biochem. Pharmacol. 2013, 85, 449–465. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.; Coombes, R.C. Estrogen receptor alpha in human breast cancer: Occurrence and significance. J. Mammary Gland Biol. Neoplasia 2000, 5, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Luparello, C.; Romanotto, R.; Tipa, A.; Sirchia, R.; Olmo, N.; López de Silanes, I.; Turnay, J.; Lizarbe, M.A.; Stewart, A.F. Midregion parathyroid hormone-related protein inhibits growth and invasion in vitro and tumorigenesis in vivo of human breast cancer cells. J. Bone Miner. Res. 2001, 16, 2173–2181. [Google Scholar] [CrossRef] [PubMed]
- Pucci-Minafra, I.; Fontana, S.; Cancemi, P.; Alaimo, G.; Minafra, S. Proteomic Patterns of Cultured Breast Cancer Cells and Epithelial Mammary Cells. Ann. N. Y. Acad. Sci. 2006, 963, 122–139. [Google Scholar] [CrossRef] [Green Version]
- Vigetti, D.; Passi, A. Hyaluronan synthases posttranslational regulation in cancer. Adv. Cancer Res. 2014, 123, 95–119. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holliday, D.L.; Speirs, V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011, 13, 215. [Google Scholar] [CrossRef] [Green Version]
- Guasti, L.; Squizzato, A.; Moretto, P.; Vigetti, D.; Ageno, W.; Dentali, F.; Maresca, A.M.; Campiotti, L.; Grandi, A.M.; Passi, A. In vitro effects of Apixaban on 5 different cancer cell lines. PLoS ONE 2017, 12, e0185035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, K.; Gras, C.; Capin, G.; Gimber, N.; Lehmann, M.; Mohd, S.; Puchkov, D.; Rödiger, M.; Wilhelmi, I.; Daumke, O.; et al. A SEPT1-based scaffold is required for Golgi integrity and function. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef] [Green Version]
- Vigetti, D.; Deleonibus, S.; Moretto, P.; Bowen, T.; Fischer, J.W.; Grandoch, M.; Oberhuber, A.; Love, D.C.; Hanover, J.A.; Cinquetti, R.; et al. Natural antisense transcript for hyaluronan synthase 2 (HAS2-AS1) induces transcription of HAS2 via protein O-GlcNAcylation. J. Biol. Chem. 2014, 289, 28816–28826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caon, I.; Bartolini, B.; Moretto, P.; Parnigoni, A.; Caravà, E.; Vitale, D.L.; Alaniz, L.; Viola, M.; Karousou, E.; De Luca, G.; et al. Sirtuin 1 reduces hyaluronan synthase 2 expression by inhibiting nuclear translocation of NF-kB and expression of the long-non coding RNA HAS2-AS1. J. Biol. Chem. 2020, jbc.RA119.011982. [Google Scholar] [CrossRef] [Green Version]
- Kultti, A.; Pasonen-Seppänen, S.; Jauhiainen, M.; Rilla, K.J.; Kärnä, R.; Pyöriä, E.; Tammi, R.H.; Tammi, M.I. 4-Methylumbelliferone inhibits hyaluronan synthesis by depletion of cellular UDP-glucuronic acid and downregulation of hyaluronan synthase 2 and 3. Exp. Cell Res. 2009, 315, 1914–1923. [Google Scholar] [CrossRef] [PubMed]
- Velesiotis, C.; Vasileiou, S.; Vynios, D.H. A guide to hyaluronan and related enzymes in breast cancer: Biological significance and diagnostic value. FEBS J. 2019, 286, 3057–3074. [Google Scholar] [CrossRef]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, P.; Weaver, V.M.; Werb, Z. The extracellular matrix: A dynamic niche in cancer progression. J. Cell Biol. 2012, 196, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Karousou, E.; D’Angelo, M.L.; Kouvidi, K.; Vigetti, D.; Viola, M.; Nikitovic, D.; De Luca, G.; Passi, A. Collagen VI and hyaluronan: The common role in breast cancer. Biomed. Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourguignon, L.Y.W.; Shiina, M.; Li, J.-J. Hyaluronan–CD44 Interaction Promotes Oncogenic Signaling, microRNA Functions, Chemoresistance, and Radiation Resistance in Cancer Stem Cells Leading to Tumor Progression. Adv. Cancer Res. 2014, 123, 255–275. [Google Scholar] [PubMed] [Green Version]
- Misra, S.; Hascall, V.C.; Markwald, R.R.; Ghatak, S. Interactions between Hyaluronan and Its Receptors (CD44, RHAMM) Regulate the Activities of Inflammation and Cancer. Front. Immunol. 2015, 6, 201. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, J.B.; El-Ashry, D.; Turley, E.A. Hyaluronan, Cancer-Associated Fibroblasts and the Tumor Microenvironment in Malignant Progression. Front. Cell Dev. Biol. 2018, 6, 1–13. [Google Scholar] [CrossRef]
- Kalluri, R.; Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Alkasalias, T.; Moyano-Galceran, L.; Arsenian-Henriksson, M.; Lehti, K. Fibroblasts in the tumor microenvironment: Shield or spear? Int. J. Mol. Sci. 2018, 19, 1532. [Google Scholar] [CrossRef] [Green Version]
- Albrengues, J.; Bertero, T.; Grasset, E.; Bonan, S.; Maiel, M.; Bourget, I.; Philippe, C.; Herraiz Serrano, C.; Benamar, S.; Croce, O.; et al. Epigenetic switch drives the conversion of fibroblasts into proinvasive cancer-associated fibroblasts. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigetti, D.; Viola, M.; Karousou, E.; Deleonibus, S.; Karamanou, K.; De Luca, G.; Passi, A. Epigenetics in extracellular matrix remodeling and hyaluronan metabolism. FEBS J. 2014, 281, 4980–4992. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Azpurua, J.; Hine, C.; Vaidya, A.; Myakishev-Rempel, M.; Ablaeva, J.; Mao, Z.; Nevo, E.; Gorbunova, V.; Seluanov, A. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 2013, 499, 346–349. [Google Scholar] [CrossRef] [Green Version]
- Bohaumilitzky, L.; Huber, A.-K.; Stork, E.M.; Wengert, S.; Woelfl, F.; Boehm, H. A Trickster in Disguise: Hyaluronan’s Ambivalent Roles in the Matrix. Front. Oncol. 2017, 7, 242. [Google Scholar] [CrossRef]
- Wu, M.; Cao, M.; He, Y.; Liu, Y.; Yang, C.; Du, Y.; Wang, W.; Gao, F. A novel role of low molecular weight hyaluronan in breast cancer metastasis. FASEB J. 2015, 29, 1290–1298. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Azpurua, J.; Ke, Z.; Augereau, A.; Zhang, Z.D.; Vijg, J.; Gladyshev, V.N.; Gorbunova, V.; Seluanov, A. INK4 locus of the tumor-resistant rodent, the naked mole rat, expresses a functional p15/p16 hybrid isoform. Proc. Natl. Acad. Sci. USA 2015, 112, 1053–1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bharadwaj, A.G.; Goodrich, N.P.; McAtee, C.O.; Haferbier, K.; Oakley, G.G.; Wahl, J.K.; Simpson, M.A. Hyaluronan suppresses prostate tumor cell proliferation through diminished expression of N-cadherin and aberrant growth factor receptor signaling. Exp. Cell Res. 2011, 317, 1214–1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camenisch, T.D.; Spicer, A.P.; Brehm-Gibson, T.; Biesterfeldt, J.; Augustine, M.L.; Calabro, A.; Kubalak, S.; Klewer, S.E.; McDonald, J.A. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J. Clin. Investig. 2000, 106, 349–360. [Google Scholar] [CrossRef] [Green Version]
- Tammi, R.H.; Passi, A.G.; Rilla, K.; Karousou, E.; Vigetti, D.; Makkonen, K.; Tammi, M.I. Transcriptional and post-translational regulation of hyaluronan synthesis. FEBS J. 2011, 278, 1419–1428. [Google Scholar] [CrossRef]
- Tammi, M.I.; Oikari, S.; Pasonen-Seppänen, S.; Rilla, K.; Auvinen, P.; Tammi, R.H. Activated hyaluronan metabolism in the tumor matrix—Causes and consequences. Matrix Biol. 2018. [Google Scholar] [CrossRef]
- Karousou, E.; Kamiryo, M.; Skandalis, S.S.; Ruusala, A.; Asteriou, T.; Passi, A.; Yamashita, H.; Hellman, U.; Heldin, C.-H.; Heldin, P. The activity of hyaluronan synthase 2 is regulated by dimerization and ubiquitination. J. Biol. Chem. 2010, 285, 23647–23654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigetti, D.; Clerici, M.; Deleonibus, S.; Karousou, E.; Viola, M.; Moretto, P.; Heldin, P.; Hascall, V.C.; De Luca, G.; Passi, A. Hyaluronan Synthesis Is Inhibited by Adenosine Monophosphate-activated Protein Kinase through the Regulation of HAS2 Activity in Human Aortic Smooth Muscle Cells. J. Biol. Chem. 2011, 286, 7917–7924. [Google Scholar] [CrossRef] [Green Version]
- Karousou, E.; Stachtea, X.; Moretto, P.; Viola, M.; Vigetti, D.; D’Angelo, M.L.; Raio, L.; Ghezzi, F.; Pallotti, F.; De Luca, G.; et al. New insights into the pathobiology of Down syndrome—Hyaluronan synthase-2 overexpression is regulated by collagen VI α2 chain. FEBS J. 2013, 280, 2418–2430. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, T.B.; Thiel, D.; Dummer, R.; Schadendorf, D.; Eichmüller, S. SEREX identification of new tumour-associated antigens in cutaneous T-cell lymphoma. Br. J. Dermatol. 2004, 150, 252–258. [Google Scholar] [CrossRef]
- Witkos, T.M.; Lowe, M. The golgin family of coiled-coil tethering proteins. Front. Cell Dev. Biol. 2016, 3, 86. [Google Scholar] [CrossRef] [Green Version]
- Cancino, J.; Jung, J.E.; Luini, A. Regulation of Golgi signaling and trafficking by the KDEL receptor. Histochem. Cell Biol. 2013, 140, 395–405. [Google Scholar] [CrossRef]
- Ailion, M.; Hannemann, M.; Dalton, S.; Pappas, A.; Watanabe, S.; Hegermann, J.; Liu, Q.; Han, H.F.; Gu, M.; Goulding, M.Q.; et al. Two Rab2 interactors regulate dense-core vesicle maturation. Neuron 2014, 82, 167–180. [Google Scholar] [CrossRef] [Green Version]
- Udabage, L.; Brownlee, G.R.; Nilsson, S.K.; Brown, T.J. The over-expression of HAS2, Hyal-2 and CD44 is implicated in the invasiveness of breast cancer. Exp. Cell Res. 2005, 310, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Evanko, S.P.; Angello, J.C.; Wight, T.N. Formation of hyaluronan- and versican-rich pericellular matrix is required for proliferation and migration of vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 1999, 19, 1004–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akiyama, Y.; Jung, S.; Salhia, B.; Lee, S.; Hubbard, S.; Taylor, M.; Mainprize, T.; Akaishi, K.; Van Furth, W.; Rutka, J.T. Hyaluronate receptors mediating glioma cell migration and proliferation. J. Neurooncol. 2001, 53, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Vanneste, M.; Hanoux, V.; Bouakka, M.; Bonnamy, P.J. Hyaluronate synthase-2 overexpression alters estrogen dependence and induces histone deacetylase inhibitor-like effects on ER-driven genes in MCF7 breast tumor cells. Mol. Cell. Endocrinol. 2017, 444, 48–58. [Google Scholar] [CrossRef]
- Bouris, P.; Skandalis, S.S.; Piperigkou, Z.; Afratis, N.; Karamanou, K.; Aletras, A.J.; Moustakas, A.; Theocharis, A.D.; Karamanos, N.K. Estrogen receptor alpha mediates epithelial to mesenchymal transition, expression of specific matrix effectors and functional properties of breast cancer cells. Matrix Biol. 2015, 43, 42–60. [Google Scholar] [CrossRef]
- Viola, M.; Brüggemann, K.; Karousou, E.; Caon, I.; Caravà, E.; Vigetti, D.; Greve, B.; Stock, C.; De Luca, G.; Passi, A.; et al. MDA-MB-231 breast cancer cell viability, motility and matrix adhesion are regulated by a complex interplay of heparan sulfate, chondroitin−/dermatan sulfate and hyaluronan biosynthesis. Glycoconj. J. 2017, 34. [Google Scholar] [CrossRef] [PubMed]
- Basiricò, L.; Bini, L.; Fontana, S.; Pallini, V.; Minafra, S.; Pucci-Minafra, I. Proteome analysis of breast cancer cells (8701-BC) cultured from primary ductal infiltrating carcinoma: Relation to correspondent breast tissues. Breast Cancer Res. 2000, 2, 1–23. [Google Scholar] [CrossRef]
- Minafra, S.; Morello, V.; Glorioso, F.; La Fiura, A.M.; Tomasino, R.M.; Feo, S.; McIntosh, D.; Woolley, D.E. A new cell line (8701-BC) from primary ductal infiltrating carcinoma of human breast. Br. J. Cancer 1989, 60, 185–192. [Google Scholar] [CrossRef] [Green Version]
- Stoppini, M.; Mangione, P.; Monti, M.; Giorgetti, S.; Marchese, L.; Arcidiaco, P.; Verga, L.; Segagni, S.; Pucci, P.; Merlini, G.; et al. Proteomics of β2-microglobulin amyloid fibrils. Biochim. Biophys. Acta—Proteins Proteom. 2005, 1753, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Gebäck, T.; Schulz, M.M.P.; Koumoutsakos, P.; Detmar, M. TScratch: A novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques 2009, 46, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Karousou, E.G.; Militsopoulou, M.; Porta, G.; De Luca, G.; Hascall, V.C.; Passi, A. Polyacrylamide gel electrophoresis of fluorophore-labeled hyaluronan and chondroitin sulfate disaccharides: Application to the analysis in cells and tissues. Electrophoresis 2004, 25, 2919–2925. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caon, I.; D’Angelo, M.L.; Bartolini, B.; Caravà, E.; Parnigoni, A.; Contino, F.; Cancemi, P.; Moretto, P.; Karamanos, N.K.; Passi, A.; et al. The Secreted Protein C10orf118 Is a New Regulator of Hyaluronan Synthesis Involved in Tumour-Stroma Cross-Talk. Cancers 2021, 13, 1105. https://doi.org/10.3390/cancers13051105
Caon I, D’Angelo ML, Bartolini B, Caravà E, Parnigoni A, Contino F, Cancemi P, Moretto P, Karamanos NK, Passi A, et al. The Secreted Protein C10orf118 Is a New Regulator of Hyaluronan Synthesis Involved in Tumour-Stroma Cross-Talk. Cancers. 2021; 13(5):1105. https://doi.org/10.3390/cancers13051105
Chicago/Turabian StyleCaon, Ilaria, Maria Luisa D’Angelo, Barbara Bartolini, Elena Caravà, Arianna Parnigoni, Flavia Contino, Patrizia Cancemi, Paola Moretto, Nikos K. Karamanos, Alberto Passi, and et al. 2021. "The Secreted Protein C10orf118 Is a New Regulator of Hyaluronan Synthesis Involved in Tumour-Stroma Cross-Talk" Cancers 13, no. 5: 1105. https://doi.org/10.3390/cancers13051105