Skin Lesion Classification Based on Surface Fractal Dimensions and Statistical Color Cluster Features Using an Ensemble of Machine Learning Techniques
Abstract
:Simple Summary
Abstract
1. Introduction
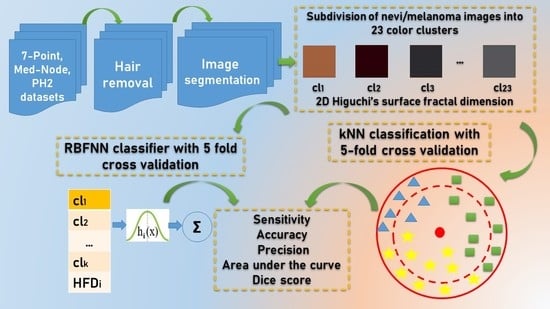
- (i)
- Determining the representative average percentage of color areas of skin lesions for each considered dataset.
- (ii)
- Proposing a descriptor for the investigation of the skin surface fractal dimensions for the channels in RGB color images, i.e., 2D Higuchi fractal dimension as an objective quantitative.
- (iii)
- Two distinctive machine learning classifiers, namely a kNN-CV algorithm and a RBFNN approach as a non-linear classifier, are implemented to generate the prediction. A dynamic partitioning of data is carried out using the 5-fold cross validation method (CV). These machine learning classifiers belong to different classification paradigms.
2. Related Works
3. Proposed Methodology
3.1. Analysis of Relevant Color Distribution in Melanocytic Lesion Images and Color Clusters Selection
3.2. Higuchi’s Surface Fractal Dimension (HFD)
3.3. K-Nearest Neighbor (kNN) with 5-Fold cross Validation (kNN-CV)
3.4. RBFNN Classifier
3.5. Dataset Description
4. Results and Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Australian Government. Melanoma of the Skin Statistics. Available online: https://melanoma.canceraustralia.gov.au/statistics (accessed on 29 March 2019).
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Silverberg, E.; Boring, C.C.; Squires, T.S. Cancer Statistics. CA Cancer J. Clin. 1990, 40, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, J.; Daneshvar, S.; Argenziano, G.; Hamarneh, G. Seven-Point checklist and skin lesion classification using multitask multimodal neural nets. IEEE J. Biomed. Health Inform. 2019, 23, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, S.; Solomon, A. A series of melanomas smaller than 4mm and implications for the ABCDE rule. J. Eur. Acad. Dermatol. Venereol. 2007, 21, 929–934. [Google Scholar] [CrossRef]
- Turani, Z.; Fatemizadeh, E.; Blumetti, T.; Daveluy, S.; Moraes, A.F.; Chen, W.; Mehregan, D.; Andersen, P.; Nisiriavanaki, M. Optical radiomic signatures derived from optical coherence tomography im- ages to improve identification of melanoma. Cancer Res. 2019, 79, 2021–2030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Codella, N.; Nguyen, Q.-B.; Pankanti, S.; Gutman, D.; Helba, B.; Halpern, A.; Smith, J. Deep learning ensembles for melanoma recognition in dermoscopy images. IBM J. Res. Dev. 2017, 61, 5:1–5:15. [Google Scholar] [CrossRef] [Green Version]
- Codella, N.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.; Dusza, S.; Kalloo, A.; Liopyris, K.; Mishra, N.K.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 International Symposium on Biomedical Imaging (ISBI), hosted by the International Skin Imaging Collaboration (ISIC). In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 168–172. [Google Scholar]
- Marchetti, M.; Codella, N.; Dusza, S.; Gutman, D.; Helba, B.; Kalloo, A.; Mishra, N.; Carrera, C.; Celebi, E.; DeFazio, J.; et al. Results of the 2016 International Skin Imaging Collaboration International Symposium on Biomedical Imaging Challenge: Comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images. J. Am. Acad. Dermatol. 2018, 78, 270–277. [Google Scholar] [CrossRef] [Green Version]
- Kasmi, R.; Mokrani, K. Classification of malignant melanoma and benign skin lesions: Implementation of automatic ABCD rule. IET Image Process. 2016, 10, 448–455. [Google Scholar] [CrossRef]
- Damian, F.A.; Moldovanu, S.; Dey, N.; Ashour, A.S.; Moraru, L. Feature Selection of Non-Dermoscopic Skin Lesion Images for Nevus and Melanoma Classification. Computation 2020, 8, 41. [Google Scholar] [CrossRef]
- Barata, C.; Celebi, M.E.; Marques, J.S. Towards a robust analysis of dermoscopy images acquired under different conditions. In Dermoscopy Image Analysis; Celebi, M.E., Mendonca, T., Marques, J.S., Eds.; CRC Press: Boca Raton, FL, USA, 2015; pp. 1–22. [Google Scholar]
- Barata, C.; Celebi, M.E.; Marques, J.S. A survey of feature extraction in dermoscopy image analysis of skin cancer. IEEE J. Biomed. Health Inform. 2019, 23, 1096–1109. [Google Scholar] [CrossRef]
- Oliveira, R.B.; Papa, J.P.; Pereira, A.S.; Tavares, J.M.R.S. Computational methods for pigmented skin lesion classification in images: Review and future trends. Neural. Comput. Appl. 2018, 29, 613–636. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.Q.; Hussain, A.; Rehman, S.U.; Khan, U.; Maqsood, M.; Mehmood, K.; Khan, M.A. Classification of Melanoma and Nevus in Digital Images for Diagnosis of Skin Cancer. IEEE Access 2019, 7, 90132–90144. [Google Scholar] [CrossRef]
- Hintz-Madsen, M.; Hansen, L.K.; Larsen, J.; Drzewiecki, K.T. A Probabilistic Neural Network Framework for the Detection of Malignant Melanoma. In Artificial Neural Networks in Cancer Diagnosis, Prognosis, and Patient Management, 1st ed.; Naguib, R.N.G., Sherbet, G.A., Eds.; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Piantanelli, A.; Maponi, P.; Scalise, L.; Serresi, S.; Cialabrini, A.; Basso, A. Fractal characterisation of boundary irregularity in skin pigmented lesions. Med Biol. Eng. Comput. 2005, 43, 436–442. [Google Scholar] [CrossRef]
- Spasic, S.; Kesic, S.; Kalauzi, A.; Saponjic, J. Different anaesthesia in rat induces distinct inter-structure brain dynamic detected by Higuchi fractal dimension. Fractals 2011, 19, 113–123. [Google Scholar] [CrossRef]
- Ali, A.-R.; Li, J.; Yang, G.; O’Shea, S.J. A machine learning approach to automatic detection of irregularity in skin lesion border using dermoscopic images. PeerJ Comput. Sci. 2020, 6, e268. [Google Scholar] [CrossRef]
- Spasic, S. On 2D generalization of Higuchi’s fractal dimension. Chaos Solitons Fractals 2014, 69, 179–187. [Google Scholar] [CrossRef]
- Klonowski, W.; Stepien, P.; Stepien, R.; Sedivy, R.; Ahammer, H.; Spasic, S. Analysis of Anal Intraepithelial Neoplasia images using 1D and 2D Higuchi’s fractal dimension methods. Fractals 2018, 26, 1850021. [Google Scholar] [CrossRef]
- Moldovanu, S.; Obreja, C.-D.; Biswas, K.C.; Moraru, L. Towards Accurate Diagnosis of Skin Lesions Using Feedforward Back Propagation Neural Networks. Diagnostics 2021, 11, 936. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.P.; Czakon, J.; Kaczmarek, K.A.; Pyskir, A.; Tarasiewicz, P.; Kunwar, S.; Rohrbach, J.; Luo, D.; Prasad, M.; Fleer, S.; et al. Deep Learning for Understanding Satellite Imagery: An Experimental Survey. Front. Artif. Intell. 2020, 3, 534696. [Google Scholar] [CrossRef]
- Fiorucci, M.; Khoroshiltseva, M.; Pontil, M.; Traviglia, A.; Del Bue, A.; James, S. Machine Learning for Cultural Heritage: A Survey. Pattern Recognit. Lett. 2020, 133, 102–108. [Google Scholar] [CrossRef]
- Rundo, L.; Han, C.; Nagano, Y.; Zhang, J.; Hataya, R.; Militello, C.; Tangherloni, A.; Nobile, M.; Ferretti, C.; Besozzi, D.; et al. USE-Net, Incorporating Squeeze-and-Excitation blocks into U-Net for prostate zonal segmentation of multi-institutional MRI datasets. Neurocomputin 2019, 365, 31–43. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Liu, Q.; Wang, Y. Road extraction by deep residual u-net. IEEE Geosci. Remote Sens. Lett. 2018, 15, 749–753. [Google Scholar] [CrossRef] [Green Version]
- Bardis, M.; Houshyar, R.; Chantaduly, C.; Tran-Harding, K.; Ushinsky, A.; Chahine, C.; Rupasinghe, M.; Chow, D.; Chang, P. Segmentation of the Prostate Transition Zone and Peripheral Zone on MR Images with Deep Learning. Radiol. Imaging Cancer 2021, 3, e200024. [Google Scholar] [CrossRef] [PubMed]
- Leopold, H.A.; Orchard, J.; Zelek, J.S.; Lakshminarayanan, V. PixelIBMM Augmenting the pixelICNN with batch normalization and the presentation of a fast architecture for retinal vessel segmentation. J. Imaging 2019, 5, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Li, Z.; Cao, L.; Balas, V.E.; Dey, N.; Ashour, A.S.; McCauley, P.; Shi, F. Multi-scale plantar pressure imaging data fusion incorporating improved Gaussian mixture operator and fuzzy weighted evaluation system. IEEE Sens. J. 2017, 17, 1407–1420. [Google Scholar] [CrossRef]
- Wang, D.; Li, Z.; Dey, N.; Ashour, A.S.; Moraru, L.; Biswas, A.; Shi, F. Optical pressure sensors based plantar image segmenting using an improved fully convolutional network. Optik 2019, 179, 99–114. [Google Scholar] [CrossRef]
- Almansour, E.; Jaffar, M.A. Classification of Dermoscopic skin cancer images using color and hybrid texture features. Int. J. Comput. Sci. Netw. Secur. 2016, 16, 135–139. [Google Scholar]
- Seidenari, S.; Grana, C.; Pellacani, G. Colour Clusters for Computer Diagnosis of Melanocytic Lesions. Dermatology 2007, 214, 137–143. [Google Scholar] [CrossRef]
- Li, X.; Wu, J.; Jiang, H.; Chen, E.Z.; Dong, X.; Rong, R. Skin Lesion Classification Via Combining Deep Learning Features and Clinical Criteria Representations. bioRxiv 2018. [Google Scholar] [CrossRef]
- Shi, C.-T. Signal Pattern Recognition Based on Fractal Features and Machine Learning. Appl. Sci. 2018, 8, 1327. [Google Scholar] [CrossRef] [Green Version]
- Čukić, M.; Stokić, M.; Simić, S.V.; Pokrakjak, D. The successful discrimination of depression from EEG could be attributed to proper feature extraction and not to a particular classification method. Cogn. Neurodynamics 2020, 14, 443–455. [Google Scholar] [CrossRef] [Green Version]
- Ahammer, H.; Sabathiel, N.; Reiss, M.A. Is a two-dimensional generalization of the Higuchi algorithm necessary? Chaos 2015, 25, 073104. [Google Scholar] [CrossRef]
- Bonyad, M.; Tieng, Q.; Reutens, D. Optimization of distributions differences for classification. IEEE Trans. Neural. Netw. Learn. Syst. 2019, 30, 511–523. [Google Scholar] [CrossRef] [Green Version]
- Tembusai, Z.R.; Mawengkang, H.; Zarlis, M. K-Nearest Neighbor with K-Fold Cross Validation and Analytic Hierarchy Process on Data Classification. Int. J. Adv. Data Inf. Syst. 2021, 2, 1–8. [Google Scholar] [CrossRef]
- Hashemi Fath, A.; Madanifar, F.; Abbasi, M. Implementation of multilayer perceptron (MLP) and radial basis function (RBF) neural networks to predict solution gas-oil ratio of crude oil systems. Petroleum 2020, 6, 80–91. [Google Scholar] [CrossRef]
- Alexandridis, A.; Chondrodima, E. A medical diagnostic tool based on radial basis function classifiers and evolutionary simulated annealing. J. Biomed. Informatics. 2014, 49, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, T.; Ferreira, P.M.; Marques, J.S.; Marcal, A.R.; Rozeira, J. PH2—A dermoscopic image database for research and benchmarking. In Proceedings of the 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 5437–5440. [Google Scholar]
- Giotis, I.; Molders, N.; Land, S.; Biehl, M.; Jonkman, M.F.; Petkov, N. MED-NODE: A computer-assisted melanoma diagnosis system using non-dermoscopic images. Expert Syst. Appl. 2015, 42, 6578–6585. [Google Scholar] [CrossRef]
- Paolino, G.; Panetta, C.; Cota, C.; Didona, D.; Moliterni, E.; Di Mattia, C.; De Vita, G.; Bottoni, U.; Donati, P.; Calvieri, S. Vitamin D receptor immunohistochemistry variability in sun-exposed and non-sun-exposed melanomas. Melanoma Res. 2017, 27, 17–23. [Google Scholar] [CrossRef]
- Paolino, G.; Moliterni, E.; Didona, D.; Garelli, V.; Corsetti, P.; Lopez, T.; Richetta, A.G.; Cantisani, C.; Bottoni, U.; Calvieri, S. Clinicopathological features, vitamin D serological levels and prognosis in cutaneous melanoma of shield-sites: An update. Med. Oncol. 2015, 32, 451. [Google Scholar] [CrossRef]
- Mercuri, S.R.; Rizzo, N.; Bellinzona, F.; Pampena, R.; Brianti, P.; Moffa, G.; Colombo Flink, L.; Bearzi, P.; Longo, C.; Paolino, G. Digital ex-vivo confocal imaging for fast Mohs surgery in nonmelanoma skin cancers: An emerging technique in dermatologic surgery. Dermatol. Ther. 2019, 32, e13127. [Google Scholar] [CrossRef]
- Russo, T.; Piccolo, V.; Lallas, A.; Jacomel, J.; Moscarella, E.; Alfano, R.; Argenziano, G. Dermoscopy of Malignant Skin Tumours: What’s New? Dermatology 2017, 233, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Skvara, H.; Teban, L.; Fiebiger, M.; Binder, M.; Kittler, H. Limitations of Dermoscopy in the Recognition of Melanoma. Arch. Dermatol. 2005, 141, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, B.G.; Goldstein, A.O. Diagnosis and management of malignant melanoma. Am. Fam. Physician 2001, 63, 1359–1368, Erratum in 2001, 64, 1682. [Google Scholar]
- Nasiri, S.; Helsper, J.; Jung, M.; Fathi, M. DePicT Melanoma Deep-CLASS: A deep convolutional neural networks approach to classify skin lesion images. BMC Bioinform. 2020, 21, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavitha, J.C.; Suruliandi, A.; Nagarajan, D.; Nadu, T. Melanoma Detection in Dermoscopic Images using Global and Local Feature Extraction. Int. J. Multimed. Ubiquit. Eng. 2017, 12, 19–28. [Google Scholar] [CrossRef]
- Al-masni, M.A.; Kim, D.-H.; Kim, T.-S. Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification. Comput. Methods Programs Biomed. 2020, 190, 105351. [Google Scholar] [CrossRef]
- Seeja, R.D.; Suresh, A. Deep learning-based skin lesion segmentation and classification of melanoma using support vector machine (SVM). Asian Pac. J. Cancer Preven 2019, 20, 1555–1561. [Google Scholar]
- Khan, M.A.; Sharif, M.; Akram, T.; Bukhari, S.A.C.; Nayak, R.S. Developed Newton-Raphson based deep features selection framework for skin lesion recognition. Pattern Recognit. Lett. 2020, 129, 293–303. [Google Scholar] [CrossRef]










| Dataset | Color Cluster Features | 2D Higuchi’s Surface Fractal |
|---|---|---|
| 7-Point | 15 average percentage of color areas: cl1, cl2, cl3, cl4, cl5, cl6, cl9, cl10, cl11, cl12, cl13 cl14, cl15, cl20, cl23 5475 features | 365 features |
| Med-Node | 10 average percentage of color areas: cl5, cl6, cl7, cl10, cl11, cl14, cl15, cl18, cl20, cl21 1400 features | 140 features |
| PH2 | 9 average percentage of color areas: cl1, cl3, cl4, cl7, cl10, cl15, cl20, cl21, cl23 1080 features | 120 features |
| Dataset | Sensitivity (%) | Accuracy (%) | Precision (%) | AUC | Dice Score |
|---|---|---|---|---|---|
| 7-Point | 80.77 | 71.43 | 73.26 | 0.6948 | 0.7683 |
| Med-Node | 30.19 | 64.23 | 57.14 | 0.6423 | 0.3951 |
| PH2 | 83.33 | 79.38 | 62.50 | 0.8047 | 0.7143 |
| Dataset | RBFNN Inputs | Sensitivity (%) | Accuracy (%) | Precision (%) | AUC | Dice Scores | No. of Hidden Neurons | MSE |
|---|---|---|---|---|---|---|---|---|
| 7-Point | Color clusters | 97.77 | 95.12 | 94.32 | 0.9412 | 0.9603 | 50 | 0.1904 |
| Color clusters and HFD | 98.01 | 95.42 | 94.44 | 0.9422 | 0.9630 | 0.0924 | ||
| Med-Node | Color clusters | 96.22 | 94.12 | 88.61 | 0.9550 | 0.9333 | 50 | 0.1789 |
| Color clusters and HFD | 96.42 | 94.71 | 87.50 | 0.9588 | 0.9396 | 0.1662 | ||
| PH2 | Color clusters | 1.00 | 94.17 | 85.03 | 0.9553 | 0.9195 | 50 | 0.1372 |
| Color clusters and HFD | 1.00 | 94.88 | 85.62 | 0.9685 | 0.9211 | 0.1128 |
| Authors | Accuracy (%) and Details |
|---|---|
| Nasiri et al. [50] | 64% (for 1st test: kNN (300, 100) and spot features) |
| 67% (2nd test: kNN (1346, 450) and spot features) | |
| Kavitha et al. [51] | 78.2 (kNN and GLCM features) |
| Al-masni et al. [52] | 81.79% (Inception-ResNet-v2, ISIC 2016 dataset) |
| 81.57% (ResNet-50, ISIC 2017 dataset) | |
| 89.29% (ResNet-50, ISIC 2018 dataset) | |
| Seeja & Suresh [53] | 79.26% (kNN, LBP and Edge histograms, HOG, Gabor filter) |
| Khan et al. [54] | 94.50% (Neural Network/Feed Forward/sigmoid function/3 hidden layers, ISBI2016 dataset, 70:30 training and testing). |
| 94.20% (Neural Network/Feed Forward/sigmoid function/3 hidden layers, ISBI2017 dataset, 70:30 training and testing). | |
| Proposed kNN-CV | 71.43% (7-Point dataset); 64.23% (Med-Node dataset) and 79.38% (PH2 dataset) for 2D Higuchi’s surface fractal features |
| Proposed (RBFNN—color clusters and HFD) | 95.42% (7-Point dataset) |
| 94.71% (Med-Node dataset) | |
| 94.88% (PH2 dataset) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moldovanu, S.; Damian Michis, F.A.; Biswas, K.C.; Culea-Florescu, A.; Moraru, L. Skin Lesion Classification Based on Surface Fractal Dimensions and Statistical Color Cluster Features Using an Ensemble of Machine Learning Techniques. Cancers 2021, 13, 5256. https://doi.org/10.3390/cancers13215256
Moldovanu S, Damian Michis FA, Biswas KC, Culea-Florescu A, Moraru L. Skin Lesion Classification Based on Surface Fractal Dimensions and Statistical Color Cluster Features Using an Ensemble of Machine Learning Techniques. Cancers. 2021; 13(21):5256. https://doi.org/10.3390/cancers13215256
Chicago/Turabian StyleMoldovanu, Simona, Felicia Anisoara Damian Michis, Keka C. Biswas, Anisia Culea-Florescu, and Luminita Moraru. 2021. "Skin Lesion Classification Based on Surface Fractal Dimensions and Statistical Color Cluster Features Using an Ensemble of Machine Learning Techniques" Cancers 13, no. 21: 5256. https://doi.org/10.3390/cancers13215256