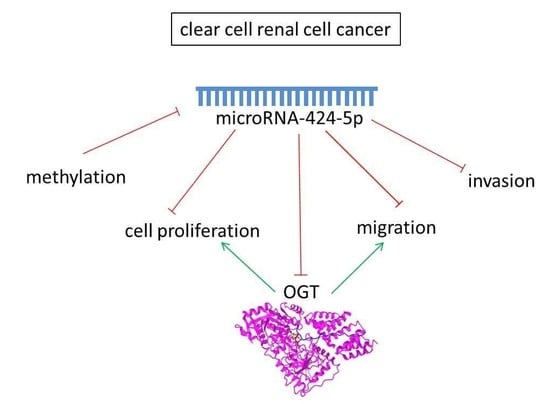
MiRNA-424-5p Suppresses Proliferation, Migration, and Invasion of Clear Cell Renal Cell Carcinoma and Attenuates Expression of O-GlcNAc-Transferase
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Prediction of miRNA Targets
2.3. Cell Transfection of Pre-miR Constructs
2.4. Cell Proliferation Assay
2.5. Cell Migration and Invasion Assays
2.6. Western Blot Analysis
2.7. 5′ Aza Treatment of Cells
2.8. Lentiviral Infection of OGT shRNA
2.9. Luciferase Assay
2.10. Statistical Analysis
3. Results
3.1. Overexpression of miR-424-5p Inhibits In Vitro Proliferation, Migration, and Invasion of 786-O and Caki-1 Cells
3.2. miR-424-5p Directly Regulates OGT by Targeting its 3′UTR
3.3. Treatment with 5′Aza Reduces Proliferation, Migration, and Invasion with Altered Expression of miR-424-5p and OGT in ccRCC Cells
3.4. OGT Lentiviral Knockdown Suppresses ccRCC Cell Proliferation and Migration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal Cell Carcinoma. Nat. Rev. Dis. Primers 2017, 3, 17009. [Google Scholar] [CrossRef] [PubMed]
- Cairns, P. Renal Cell Carcinoma. Cancer Biomark. 2011, 9, 461–473. [Google Scholar] [CrossRef]
- Motzer, R.J.; Bacik, J.; Mariani, T.; Russo, P.; Mazumdar, M.; Reuter, V. Treatment Outcome and Survival Associated With Metastatic Renal Cell Carcinoma of Non–Clear-Cell Histology. J. Clin. Oncol. 2002, 20, 2376–2381. [Google Scholar] [CrossRef]
- Keegan, K.A.; Schupp, C.W.; Chamie, K.; Hellenthal, N.J.; Evans, C.P.; Koppie, T.M. Histopathology of Surgically Treated Renal Cell Carcinoma: Survival Differences by Subtype and Stage. J. Urol. 2012, 188, 391–397. [Google Scholar] [CrossRef] [Green Version]
- Bekelman, J.E.; Rumble, R.B.; Chen, R.C.; Pisansky, T.M.; Finelli, A.; Feifer, A.; Nguyen, P.L.; Loblaw, D.A.; Tagawa, S.T.; Gillessen, S.; et al. Clinically Localized Prostate Cancer: ASCO Clinical Practice Guideline Endorsement of an American Urological Association/American Society for Radiation Oncology/Society of Urologic Oncology Guideline. J. Clin. Oncol. 2018, 36, 3251–3258. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Shi, X.; Wang, C.; Jin, P.; Ma, F. Transcription Factor and MiRNA Interplays Can Manifest the Survival of CcRCC Patients. Cancers 2019, 11, 1668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, L.; Wang, Z.; Chen, Q.; Li, Y.; He, H.; Hsieh, J.J.; Xue, S.; Wu, Z.; Liu, B.; Tang, H.; et al. Prognostic Value of a Long Non-Coding RNA Signature in Localized Clear Cell Renal Cell Carcinoma. Eur. Urol. 2018, 74, 756–763. [Google Scholar] [CrossRef]
- Gowrishankar, B.; Ibragimova, I.; Zhou, Y.; Slifker, M.J.; Devarajan, K.; Al-Saleem, T.; Uzzo, R.G.; Cairns, P. MicroRNA Expression Signatures of Stage, Grade, and Progression in Clear Cell RCC. Cancer Biol. Ther. 2014, 15, 329–341. [Google Scholar] [CrossRef] [Green Version]
- Moynihan, M.J.; Sullivan, T.B.; Burks, E.; Schober, J.; Calabrese, M.; Fredrick, A.; Kalantzakos, T.; Warrick, J.; Canes, D.; Raman, J.D.; et al. MicroRNA Profile in Stage I Clear Cell Renal Cell Carcinoma Predicts Progression to Metastatic Disease. Urol. Oncol. Semin. Orig. Investig. 2020, 38, 799.e11–799.e22. [Google Scholar] [CrossRef]
- Heinzelmann, J.; Henning, B.; Sanjmyatav, J.; Posorski, N.; Steiner, T.; Wunderlich, H.; Gajda, M.R.; Junker, K. Specific MiRNA Signatures Are Associated with Metastasis and Poor Prognosis in Clear Cell Renal Cell Carcinoma. World J. Urol. 2011, 29, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Slaby, O.; Redova, M.; Poprach, A.; Nekvindova, J.; Iliev, R.; Radova, L.; Lakomy, R.; Svoboda, M.; Vyzula, R. Identification of MicroRNAs Associated with Early Relapse after Nephrectomy in Renal Cell Carcinoma Patients. Genes Chromosom. Cancer 2012, 51, 707–716. [Google Scholar] [CrossRef]
- Chen, B.; Duan, L.; Yin, G.; Tan, J.; Jiang, X. Simultaneously Expressed MiR-424 and MiR-381 Synergistically Suppress the Proliferation and Survival of Renal Cancer Cells---Cdc2 Activity Is up-Regulated by Targeting WEE1. Clinics 2013, 68, 825–833. [Google Scholar] [CrossRef]
- Liu, J.; Gu, Z.; Tang, Y.; Hao, J.; Zhang, C.; Yang, X. Tumour-Suppressive MicroRNA-424-5p Directly Targets CCNE1 as Potential Prognostic Markers in Epithelial Ovarian Cancer. Cell Cycle 2018, 17, 309–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Zhao, F.; Chen, H.; Liu, Y.; Su, J. MicroRNA-424-5p Inhibits the Proliferation, Migration, and Invasion of Nasopharyngeal Carcinoma Cells by Decreasing AKT3 Expression. Braz. J. Med. Biol. Res. 2020, 53, e9029. [Google Scholar] [CrossRef]
- Wu, J.; Yang, B.; Zhang, Y.; Feng, X.; He, B.; Xie, H.; Zhou, L.; Wu, J.; Zheng, S. MiR-424-5p Represses the Metastasis and Invasion of Intrahepatic Cholangiocarcinoma by Targeting ARK5. Int. J. Biol. Sci. 2019, 15, 1591–1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Qiu, X.-M.; Li, Q.-H.; Wang, X.-Y.; Li, L.; Xu, M.; Dong, M.; Xiao, Y.-B. MicroRNA-424 May Function as a Tumor Suppressor in Endometrial Carcinoma Cells by Targeting E2F7. Oncol. Rep. 2015, 33, 2354–2360. [Google Scholar] [CrossRef]
- Dong, J.; Wang, Q.; Li, L.; Xiao, Z. Upregulation of Long Non-Coding RNA Small Nucleolar RNA Host Gene 12 Contributes to Cell Growth and Invasion in Cervical Cancer by Acting as a Sponge for MiR-424-5p. Cell Physiol. Biochem. 2018, 45, 2086–2094. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, S.; Zhou, J.; Qian, Q. MiR-424-5p Regulates Cell Proliferation, Migration and Invasion by Targeting Doublecortin-like Kinase 1 in Basal-like Breast Cancer. Biomed. Pharmacother. 2018, 102, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Li, M.; Ouyang, Y.; Tan, Z.; Jiang, Y. MiR-424 Functions as a Tumor Suppressor in Glioma Cells and Is down-Regulated by DNA Methylation. J. Neurooncol. 2017, 133, 247–255. [Google Scholar] [CrossRef]
- Wu, C.-T.; Lin, W.-Y.; Chang, Y.-H.; Lin, P.-Y.; Chen, W.-C.; Chen, M.-F. DNMT1-Dependent Suppression of MicroRNA424 Regulates Tumor Progression in Human Bladder Cancer. Oncotarget 2015, 6, 24119–24131. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Li, Y.; Gan, Y.; Tian, R.; Wu, Q.; Shu, G.; Yin, G. Methylation-Mediated Repression of MiR-424/503 Cluster Promotes Proliferation and Migration of Ovarian Cancer Cells through Targeting the Hub Gene KIF23. Cell Cycle 2019, 18, 1601–1618. [Google Scholar] [CrossRef]
- Ghonbalani, Z.N.; Shahmohamadnejad, S.; Pasalar, P.; Khalili, E. Hypermethylated MiR-424 in Colorectal Cancer Subsequently Upregulates VEGF. J. Gastrointest. Cancer 2021. [Google Scholar] [CrossRef]
- Li, Y.; Liu, J.; Hu, W.; Zhang, Y.; Sang, J.; Li, H.; Ma, T.; Bo, Y.; Bai, T.; Guo, H.; et al. MiR-424-5p Promotes Proliferation, Migration and Invasion of Laryngeal Squamous Cell Carcinoma. OncoTargets Ther. 2019, 12, 10441–10453. [Google Scholar] [CrossRef] [Green Version]
- Dai, W.; Zhou, J.; Wang, H.; Zhang, M.; Yang, X.; Song, W. MiR-424-5p Promotes the Proliferation and Metastasis of Colorectal Cancer by Directly Targeting SCN4B. Pathol. Res. Pract. 2020, 216, 152731. [Google Scholar] [CrossRef]
- Liep, J.; Kilic, E.; Meyer, H.A.; Busch, J.; Jung, K.; Rabien, A. Cooperative Effect of MiR-141-3p and MiR-145-5p in the Regulation of Targets in Clear Cell Renal Cell Carcinoma. PLoS ONE 2016, 11, e0157801. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Hatzigeorgiou, A.G. Online Resources for MiRNA Analysis. Clin. Biochem. 2013, 46, 879–900. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting Effective MicroRNA Target Sites in Mammalian MRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-MicroT Web Server v5.0: Service Integration into MiRNA Functional Analysis Workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reczko, M.; Maragkakis, M.; Alexiou, P.; Grosse, I.; Hatzigeorgiou, A.G. Functional MicroRNA Targets in Protein Coding Sequences. Bioinformatics 2012, 28, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Rehmsmeier, M.; Steffen, P.; Höchsmann, M.; Giegerich, R. Fast and Effective Prediction of MicroRNA/Target Duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial MicroRNA Target Predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef]
- Vaiana, C.A.; Kurcon, T.; Mahal, L.K. MicroRNA-424 Predicts a Role for β-1,4 Branched Glycosylation in Cell Cycle Progression. J. Biol. Chem. 2016, 291, 1529–1537. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Jin, J.; Qiu, Z.; Liu, D.; Luo, H. Functional Analysis of O-GlcNAcylation in Cancer Metastasis. Front. Oncol. 2020, 10, 585288. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, S.; Zhang, J.; Mao, S.; Mao, W.; Zhang, W.; Guo, Y.; Wu, Y.; Wang, R.; Yan, Y.; et al. Suppressed OGT Expression Inhibits Cell Proliferation and Modulates EGFR Expression in Renal Cell Carcinoma. Cancer Manag. Res. 2019, 11, 2215–2223. [Google Scholar] [CrossRef] [Green Version]
- Kalantzakos, T.J.; Sullivan, T.B.; Sebel, L.E.; Canes, D.; Burks, E.J.; Moinzadeh, A.; Rieger-Christ, K.M. MicroRNAs MiR-15a and MiR-26a Cooperatively Regulate O-GlcNAc-Transferase to Control Proliferation in Clear Cell Renal Cell Carcinoma. Cancer Biomark. 2021, 30, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Roche, J. The Epithelial-to-Mesenchymal Transition in Cancer. Cancers 2018, 10, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brodaczewska, K.K.; Szczylik, C.; Fiedorowicz, M.; Porta, C.; Czarnecka, A.M. Choosing the Right Cell Line for Renal Cell Cancer Research. Mol. Cancer 2016, 15, 83. [Google Scholar] [CrossRef] [Green Version]
- Maudet, C.; Mano, M.; Sunkavalli, U.; Sharan, M.; Giacca, M.; Förstner, K.U.; Eulalio, A. Functional High-Throughput Screening Identifies the MiR-15 MicroRNA Family as Cellular Restriction Factors for Salmonella Infection. Nat. Commun. 2014, 5, 4718. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A Pathology Atlas of the Human Cancer Transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trinca, G.M.; Goodman, M.L.; Papachristou, E.K.; D’Santos, C.S.; Chalise, P.; Madan, R.; Slawson, C.; Hagan, C.R. O-GlcNAc-Dependent Regulation of Progesterone Receptor Function in Breast Cancer. Horm. Cancer 2018, 9, 12–21. [Google Scholar] [CrossRef]
- Ferrer, C.M.; Sodi, V.L.; Reginato, M.J. O-GlcNAcylation in Cancer Biology: Linking Metabolism and Signaling. J. Mol. Biol. 2016, 428, 3282–3294. [Google Scholar] [CrossRef] [Green Version]
- Konzman, D.; Abramowitz, L.K.; Steenackers, A.; Mukherjee, M.M.; Na, H.-J.; Hanover, J.A. O-GlcNAc: Regulator of Signaling and Epigenetics Linked to X-Linked Intellectual Disability. Front. Genet. 2020, 11, 605263. [Google Scholar] [CrossRef] [PubMed]
- Boulard, M.; Rucli, S.; Edwards, J.R.; Bestor, T.H. Methylation-Directed Glycosylation of Chromatin Factors Represses Retrotransposon Promoters. Proc. Natl. Acad. Sci. USA 2020, 117, 14292–14298. [Google Scholar] [CrossRef] [PubMed]
- Barde, I.; Rauwel, B.; Marin-Florez, R.M.; Corsinotti, A.; Laurenti, E.; Verp, S.; Offner, S.; Marquis, J.; Kapopoulou, A.; Vanicek, J.; et al. A KRAB/KAP1-MiRNA Cascade Regulates Erythropoiesis Through Stage-Specific Control of Mitophagy. Science 2013, 340, 350–353. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalantzakos, T.J.; Sullivan, T.B.; Gloria, T.; Canes, D.; Moinzadeh, A.; Rieger-Christ, K.M. MiRNA-424-5p Suppresses Proliferation, Migration, and Invasion of Clear Cell Renal Cell Carcinoma and Attenuates Expression of O-GlcNAc-Transferase. Cancers 2021, 13, 5160. https://doi.org/10.3390/cancers13205160
Kalantzakos TJ, Sullivan TB, Gloria T, Canes D, Moinzadeh A, Rieger-Christ KM. MiRNA-424-5p Suppresses Proliferation, Migration, and Invasion of Clear Cell Renal Cell Carcinoma and Attenuates Expression of O-GlcNAc-Transferase. Cancers. 2021; 13(20):5160. https://doi.org/10.3390/cancers13205160
Chicago/Turabian StyleKalantzakos, Thomas J., Travis B. Sullivan, Thales Gloria, David Canes, Alireza Moinzadeh, and Kimberly M. Rieger-Christ. 2021. "MiRNA-424-5p Suppresses Proliferation, Migration, and Invasion of Clear Cell Renal Cell Carcinoma and Attenuates Expression of O-GlcNAc-Transferase" Cancers 13, no. 20: 5160. https://doi.org/10.3390/cancers13205160