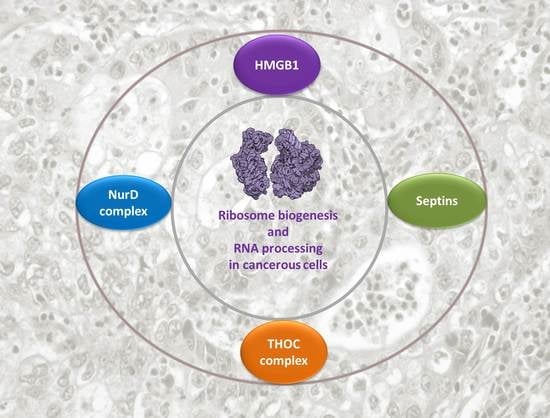
HMGB1 Protein Interactions in Prostate and Ovary Cancer Models Reveal Links to RNA Processing and Ribosome Biogenesis through NuRD, THOC and Septin Complexes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Subcellular Fractionation
2.3. Large-Scale Immunoprecipitation
2.4. Mass Spectrometry and Data Analysis
2.5. Yeast Two-Hybrid Assays
2.6. Small Scale Immunoprecipitation and Western Blot
2.7. HMGB1 Silencing
2.8. qPCR Analysis of Gene Expression
3. Results
3.1. HMGB1 Interactome in Ovary and Prostate Cancer Cell Lines
3.2. HMGB1 Interacts with Nucleosome Remodelling (NuRD) Complex Subunit RBBP7
3.3. HMGB1-Associated THOC and SEPTIN Complexes Interact between Them and with RAB11
3.4. Clinical Significance of Expression Changes of HMGB1 Interactome Components Related to RNA Processing and Nuclear Export in Prostate and Ovary Cancers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vizoso-Vázquez, Á.; Barreiro-Alonso, A.; Rico-Díaz, A.; Lamas-Maceiras, M.; Rodríguez-Belmonte, E.; Becerra, M.; González-Siso, M.; Cerdán, M. HMGB Proteins from Yeast to Human. Gene Regulation, DNA Repair and Beyond. In Old Yeasts—New Questions; InTech: Tokyo, Japan, 2017; pp. 139–165. ISBN 978-953-51-3677-4. [Google Scholar] [CrossRef]
- Barreiro-Alonso, A.; Lamas-Maceiras, M.; Rodríguez-Belmonte, E.; Vizoso-Vázquez, A.; Quindós, M.; Cerdán, M.E. High Mobility Group B Proteins, their Partners, and Other Redox Sensors in Ovarian and Prostate Cancer. Oxid Med. Cell Longev. 2016, 2016, e5845061. [Google Scholar] [CrossRef]
- Zhang, J.; Shao, S.; Han, D.; Xu, Y.; Jiao, D.; Wu, J.; Yang, F.; Ge, Y.; Shi, S.; Li, Y.; et al. High Mobility Group Box 1 Promotes the Epithelial-to-Mesenchymal Transition in Prostate Cancer PC3 Cells Via the RAGE/NF-kappaB Signaling Pathway. Int. J. Oncol. 2018, 53, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Paek, J.; Lee, M.; Nam, E.J.; Kim, S.W.; Kim, Y.T. Clinical Impact of High Mobility Group Box 1 Protein in Epithelial Ovarian Cancer. Arch. Gynecol. Obstet. 2016, 293, 645–650. [Google Scholar] [CrossRef]
- Kang, R.; Zhang, Q.; Zeh, H.J., 3rd; Lotze, M.T.; Tang, D. HMGB1 in Cancer: Good, Bad, Or both? Clin. Cancer Res. 2013, 19, 4046–4057. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, H.; Song, B.; Sun, Y.; Xu, Z.; Han, J. Silencing HMGB1 Expression by Lentivirus-Mediated Small Inter-fering RNA (siRNA) Inhibits the Proliferation and Invasion of Colorectal Cancer LoVo Cells in Vitro and in Vivo. Chin. J. Oncol. 2015, 37, 664–670. [Google Scholar]
- De Las Rivas, J.; Alonso-Lopez, D.; Arroyo, M.M. Human Interactomics: Comparative Analysis of Different Protein Interaction Resources and Construction of a Cancer Protein-Drug Bipartite Network. Adv. Protein Chem. Struct. Biol. 2018, 111, 263–282. [Google Scholar] [CrossRef] [PubMed]
- Barreiro-Alonso, A.; Cámara-Quílez, M.; Salamini-Montemurri, M.; Lamas-Maceiras, M.; Vizoso-Vázquez, A.; Rodríguez-Belmonte, E.; Quindós-Varela, M.; Martínez-Iglesias, O.; Figueroa, A.; Cerdán, M.E. Characterization of HMGB1/2 Interactome in Prostate Cancer by Yeast Two Hybrid Approach: Potential Pathobiological Implications. Cancers 2019, 11, 1729. [Google Scholar] [CrossRef] [PubMed]
- Cámara-Quílez, M.; Barreiro-Alonso, A.; Vizoso-Vázquez, A.; Rodríguez-Belmonte, E.; Quindós-Varela, M.; Lamas-Maceiras, M.; Cerdán, M.E. The HMGB1-2 Ovarian Cancer Interactome. the Role of HMGB Proteins and their In-teracting Partners MIEN1 and NOP53 in Ovary Cancer and Drug-Response. Cancers 2020, 12, 2435. [Google Scholar] [CrossRef]
- Ugrinova, I.; Pasheva, E. HMGB1 Protein: A Therapeutic Target Inside and Outside the Cell. Adv. Protein Chem. Struct. Biol. 2017, 107, 37–76. [Google Scholar] [CrossRef]
- Pardo, M.; Lang, B.; Yu, L.; Prosser, H.; Bradley, A.; Babu, M.M.; Choudhary, J. An Expanded Oct4 Interaction Net-work: Implications for Stem Cell Biology, Development, and Disease. Cell Stem Cell 2010, 6, 382–395. [Google Scholar] [CrossRef]
- Hillier, C.; Pardo, M.; Yu, L.; Bushell, E.; Sanderson, T.; Metcalf, T.; Herd, C.; Anar, B.; Rayner, J.C.; Billker, O.; et al. Landscape of the Plasmodium Interactome Reveals both Conserved and Species-Specific Functionality. Cell Rep. 2019, 28, 1635–1647.e5. [Google Scholar] [CrossRef]
- Choi, H.; Liu, G.; Mellacheruvu, D.; Tyers, M.; Gingras, A.C.; Nesvizhskii, A.I. Analyzing Protein-Protein Interac-tions from Affinity Purification-Mass Spectrometry Data with SAINT. Curr. Protoc. Bioinform. 2012. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- James, P.; Halladay, J.; Craig, E.A. Genomic Libraries and a Host Strain Designed for Highly Efficient Two-Hybrid Selection in Yeast. Genet. 1996, 144, 1425–1436. [Google Scholar] [CrossRef] [PubMed]
- Rose, M.; Botstein, D. Construction and use of Gene Fusions to lacZ (β-Galactosidase) that are Expressed in yeast. Methods Enzymol. 1983, 101, 167–180. [Google Scholar]
- Goodwin, G.H.; Sanders, C.; Johns, E.W. A New Group of Chromatin-Associated Proteins with a High Content of Acidic and Basic Amino Acids. Eur. J. Biochem. 1973, 38, 14–19. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING Database in 2017: Quality-Controlled Protein-Protein Association Networks, made Broadly Accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Kloet, S.L.; Baymaz, H.I.; Makowski, M.; Groenewold, V.; Jansen, P.W.; Berendsen, M.; Niazi, H.; Kops, G.J.; Ver-meulen, M. Towards Elucidating the Stability, Dynamics and Architecture of the Nucleosome Remodeling and Deacetylase Complex by using Quantitative Interaction Proteomics. FEBS J. 2015, 282, 1774–1785. [Google Scholar] [CrossRef]
- Lai, A.Y.; Wade, P.A. Cancer Biology and NuRD: A Multifaceted Chromatin Remodelling Complex. Nat. Rev. Cancer 2011, 11, 588–596. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.; Zhang, S.; Jiao, J.; Zhang, T.; Qu, W.; Muloye, G.M.; Kong, B.; Zhang, Q.; Cui, B. Promoter Methylation of SEPT9 as a Potential Biomarker for Early Detection of Cervical Cancer and its Overexpression Predicts Radioresistance. Clin. Epigenetics 2019, 11, 120. [Google Scholar] [CrossRef] [PubMed]
- Mostowy, S.; Cossart, P. Septins: The Fourth Component of the Cytoskeleton. Nat. Rev. Mol. Cell Biol. 2012, 13, 183–194. [Google Scholar] [CrossRef]
- Desterke, C.; Gassama-Diagne, A. Protein-Protein Interaction Analysis Highlights the Role of Septins in Membrane Enclosed Lumen and mRNA Processing. Adv. Biol. Regul. 2019, 73, e100635. [Google Scholar] [CrossRef]
- Masuda, S.; Das, R.; Cheng, H.; Hurt, E.; Dorman, N.; Reed, R. Recruitment of the Human TREX Complex to mRNA during Splicing. Genes Dev. 2005, 19, 1512–1517. [Google Scholar] [CrossRef]
- Puhringer, T.; Hohmann, U.; Fin, L.; Pacheco-Fiallos, B.; Schellhaas, U.; Brennecke, J.; Plaschka, C. Structure of the Human Core Transcription-Export Complex Reveals a Hub for Multivalent Interactions. Elife 2020, 9. [Google Scholar] [CrossRef]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Pre-iss, T.; Steinmetz, L.M.; et al. Insights into RNA Biology from an Atlas of Mammalian mRNA-Binding Proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Corkery, D.P.; Holly, A.C.; Lahsaee, S.; Dellaire, G. Connecting the Speckles: Splicing Kinases and their Role in Tumorigenesis and Treatment Response. Nucleus 2015, 6, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Vizoso-Vázquez, A.; Barreiro-Alonso, A.; González-Siso, M.I.; Rodríguez-Belmonte, E.; Lamas-Maceiras, M.; Cerdán, M.E. HMGB Proteins Involved in TOR Signaling as General Regulators of Cell Growth by Controlling Ribosome Biogenesis. Curr. Genet. 2018, 64, 1205–1213. [Google Scholar] [CrossRef]
- Hopkins, T.G.; Mura, M.; Al-Ashtal, H.A.; Lahr, R.M.; Abd-Latip, N.; Sweeney, K.; Lu, H.; Weir, J.; El-Bahrawy, M.; Steel, J.H.; et al. The RNA-Binding Protein LARP1 is a Post-Transcriptional Regulator of Survival and Tumorigenesis in Ovarian Cancer. Nucleic Acids Res. 2016, 44, 1227–1246. [Google Scholar] [CrossRef] [PubMed]
- Sloan, K.E.; Bohnsack, M.T.; Watkins, N.J. The 5S RNP Couples p53 Homeostasis to Ribosome Biogenesis and Nu-cleolar Stress. Cell. Rep. 2013, 5, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Vellky, J.E.; Ricke, E.A.; Huang, W.; Ricke, W.A. Expression, Localization, and Function of the Nucleolar Protein BOP1 in Prostate Cancer Progression. Am. J. Pathol. 2021, 191, 168–179. [Google Scholar] [CrossRef]
- Hamdane, N.; Herdman, C.; Mars, J.C.; Stefanovsky, V.; Tremblay, M.G.; Moss, T. Depletion of the Cisplatin Tar-geted HMGB-Box Factor UBF Selectively Induces p53-Independent Apoptotic Death in Transformed Cells. Oncotarget 2015, 6, 27519–27536. [Google Scholar] [CrossRef]
- Sanij, E.; Diesch, J.; Lesmana, A.; Poortinga, G.; Hein, N.; Lidgerwood, G.; Cameron, D.P.; Ellul, J.; Goodall, G.J.; Wong, L.H.; et al. A Novel Role for the Pol I Transcription Factor UBTF in Maintaining Genome Stability through the Regulation of Highly Transcribed Pol II Genes. Genome Res. 2015, 25, 201–212. [Google Scholar] [CrossRef]
- Sturm, M.; Cheng, J.; Bassler, J.; Beckmann, R.; Hurt, E. Interdependent Action of KH Domain Proteins Krr1 and Dim2 Drive the 40S Platform Assembly. Nat. Commun. 2017, 8, 2213. [Google Scholar] [CrossRef]
- Miyazawa, N.; Yoshikawa, H.; Magae, S.; Ishikawa, H.; Izumikawa, K.; Terukina, G.; Suzuki, A.; Nakamu-ra-Fujiyama, S.; Miura, Y.; Hayano, T.; et al. Human Cell Growth Regulator Ly-1 Antibody Reactive Homologue Accel-erates Processing of Preribosomal RNA. Genes Cells 2014, 19, 273–286. [Google Scholar] [CrossRef]
- Abetov, D.A.; Kiyan, V.S.; Zhylkibayev, A.A.; Sarbassova, D.A.; Alybayev, S.D.; Spooner, E.; Song, M.S.; Bersim-baev, R.I.; Sarbassov, D.D. Formation of Mammalian Preribosomes Proceeds from Intermediate to Composed State during Ribosome Maturation. J. Biol. Chem. 2019, 294, 10746–10757. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Ling, T.; Zhou, Y.; Feng, W.; Zhu, Q.; Stunnenberg, H.G.; Grummt, I.; Tao, W. The Chromatin Remodeling Complex NuRD Establishes the Poised State of rRNA Genes Characterized by Bivalent Histone Modifications and Al-tered Nucleosome Positions. Proc. Natl. Acad. Sci. USA 2012, 109, 8161–8166. [Google Scholar] [CrossRef] [PubMed]
- Heath, C.G.; Viphakone, N.; Wilson, S.A. The Role of TREX in Gene Expression and Disease. Biochem. J. 2016, 473, 2911–2935. [Google Scholar] [CrossRef]
- Chinnam, M.; Wang, Y.; Zhang, X.; Gold, D.L.; Khoury, T.; Nikitin, A.Y.; Foster, B.A.; Li, Y.; Bshara, W.; Morrison, C.D.; et al. The Thoc1 Ribonucleoprotein and Prostate Cancer Progression. J. Natl. Cancer Inst. 2014, 106, dju306. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.D.; Koch, A.; Tamura, T. THOC5, a Member of the mRNA Export Complex: A Novel Link between mRNA Export Machinery and Signal Transduction Pathways in Cell Proliferation and Differentiation. Cell. Commun. Signal 2014, 12, 3. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.D.; Saran, S.; Koch, A.; Tamura, T. MRNA Export Protein THOC5 as a Tool for Identification of Target Genes for Cancer Therapy. Cancer Lett. 2016, 373, 222–226. [Google Scholar] [CrossRef]
- Fung, K.Y.; Dai, L.; Trimble, W.S. Cell and Molecular Biology of Septins. Int. Rev. Cell. Mol. Biol. 2014, 310, 289–339. [Google Scholar] [CrossRef] [PubMed]
- Dias, A.P.; Dufu, K.; Lei, H.; Reed, R. A Role for TREX Components in the Release of Spliced mRNA from Nuclear Speckle Domains. Nat. Commun. 2010, 1, 97. [Google Scholar] [CrossRef] [PubMed]
- Gilad, R.; Meir, K.; Stein, I.; German, L.; Pikarsky, E.; Mabjeesh, N.J. High SEPT9_i1 Protein Expression is Associated with High-Grade Prostate Cancers. PLoS ONE 2015, 10, e0124251. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef] [PubMed]







| SKOV-3 | PC-3 | ||||||
|---|---|---|---|---|---|---|---|
| ID | Description | OGC | BGC | FDR | OGC | BGC | FDR |
| GO:0000445 | THO complex part of TREX | 4 | 6 | 2.04 × 10−6 | 4 | 6 | 2.64 × 10−7 |
| GO:0006396 | RNA processing | 54 | 825 | 6.15 × 10−41 | ns | ||
| GO:0000398 | mRNA splicing, via spliceosome | 35 | 284 | 1.91 × 10−33 | 14 | 284 | 6.00 × 10−12 |
| GO:0006364 | rRNA processing | 8 | 192 | 0.00024 | ns | ||
| GO:1990904 | Ribonucleoprotein complex | 39 | 770 | 5.65 × 10−25 | 13 | 770 | 8.23 × 10−7 |
| GO:0022613 | RNP complex biogenesis | 20 | 409 | 8.51 × 10−12 | 8 | 409 | 0.00038 |
| GO:0000387 | Spliceosomal snRNP assembly | 4 | 37 | 0.0010 | ns | ||
| GO:0071426 | RNP complex export from nucleus | 20 | 124 | 9.01 × 10−21 | ns | ||
| GO:0006325 | Chromatin organisation | 13 | 683 | 0.0016 | ns | ||
| GO:0034728 | Nucleosome organisation | 7 | 167 | 0.00074 | ns | ||
| GO:0040029 | Regulation of gene expression, epigenetic | ns | 6 | 251 | 0.0016 | ||
| GO:0016581 | NuRD complex | 2 | 14 | 0.0187 | 2 | 14 | 4.00 × 10−3 |
| GO:0031105 | Septin complex | 4 | 6 | 2.04 × 10−6 | 4 | 6 | 2.64 × 10−7 |
| GO:0051301 | Cell division | 10 | 483 | 0.0049 | 9 | 483 | 1.7 × 10−4 |
| GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 11 | 92 | 3.44 × 10−10 | ns | ||
| GO:0002181 | Cytoplasmic translation | 4 | 57 | 0.0044 | ns | ||
| GO:0006417 | Regulation of translation | 7 | 327 | 0.0258 | ns | ||
| IPR016491 | Septin | 8 | 13 | 1.03 × 10−11 | 8 | 13 | 3.40 × 10−14 |
| SKOV-3 | PC-3 | ||||||
|---|---|---|---|---|---|---|---|
| ID | Description | OGC | BGC | FDR | OGC | BGC | FDR |
| NUCLEUS | |||||||
| GO:0006396 | RNA processing | 119 | 825 | 6.10 × 10−75 | 100 | 825 | 3.72 × 10−59 |
| GO:0000398 | mRNA splicing, via spliceosome | 65 | 284 | 7.83 × 10−50 | 41 | 284 | 1.94 × 10−25 |
| GO:0006364 | rRNA processing | 43 | 192 | 3.72 × 10−32 | 51 | 192 | 3.54 × 10−43 |
| GO:0000470 | Maturation of LSU-rRNA | 12 | 24 | 2.69 × 10−12 | 14 | 24 | 1.87 × 10−15 |
| GO:0030490 | Maturation of SSU-rRNA | 12 | 52 | 4.08 × 10−9 | 15 | 52 | 5.52 × 10−13 |
| GO:0000460 | Maturation of 5.8S rRNA | 4 | 32 | 0.0196 | 9 | 32 | 1.21 × 10−6 |
| GO:1990904 | Ribonucleoprotein complex | 149 | 770 | 9.45 × 10−115 | 141 | 770 | 2.47 × 10−110 |
| GO:0022613 | RNP complex biogenesis | 81 | 409 | 1.37 × 10−58 | 79 | 409 | 3.94 × 10−59 |
| GO:0000387 | Spliceosomal snRNP assembly | 10 | 37 | 3.27 × 10−8 | 7 | 37 | 3.70 × 10−5 |
| GO:0051169 | Nuclear transport | 28 | 267 | 3.38 × 10−13 | 27 | 267 | 3.59 × 10−13 |
| GO:0071426 | RNP complex nuclear export | 18 | 124 | 1.37 × 10−10 | 17 | 124 | 3.49 × 10−10 |
| GO:0006325 | Chromatin organisation | 42 | 683 | 2.47 × 10−12 | 40 | 683 | 3.67 × 10−12 |
| GO:0034728 | Nucleosome organisation | 22 | 167 | 3.66 × 10−12 | 22 | 167 | 3.66 × 10−12 |
| GO:0040029 | Regulation of gene expression, epigenetic | 21 | 251 | 2.45 × 10−8 | 17 | 251 | 4.77 × 10−6 |
| GO:0016581 | NuRD complex | 6 | 14 | 3.05 × 10−6 | 5 | 14 | 4.20 × 10−5 |
| GO:0031105 | Septin complex | 4 | 6 | 6.96 × 10−5 | 4 | 6 | 5.79 × 10−5 |
| GO:0006281 | DNA repair | 19 | 491 | 0.0039 | 15 | 491 | 0.0440 |
| GO:0051301 | Cell division | 21 | 483 | 0.00036 | 24 | 483 | 4.47 × 10−6 |
| CYTOPLASM | |||||||
| GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 60 | 92 | 2.15 × 10−96 | 29 | 92 | 2.53 × 10−43 |
| GO:0002181 | Cytoplasmic translation | 27 | 57 | 6.52 × 10−39 | 13 | 57 | 1.72 × 10−17 |
| GO:0006417 | Regulation of translation | 12 | 327 | 1.39 × 10−5 | 11 | 327 | 1.31 × 10−6 |
| GO:0051301 | Cell division | 11 | 483 | 0.0025 | 12 | 483 | 6.70 × 10−6 |
| IPR016491 | Septin | 8 | 13 | 4.74 × 10−11 | 8 | 13 | 1.06 × 10−12 |
| SKOV-3 | PC-3 | ||||||
|---|---|---|---|---|---|---|---|
| ID | Description | OGC | BGC | FDR | OGC | BGC | FDR |
| GO:0005634 | Nucleus | 243 | 6892 | 7.34 × 10−55 | 226 | 6892 | 1.96 × 10−51 |
| GO:0005654 | Nucleoplasm | 192 | 3446 | 5.34 × 10−66 | 165 | 3446 | 1.45 × 10−50 |
| GO:0005730 | Nucleolus | 79 | 926 | 2.50 × 10−33 | 88 | 926 | 9.18 × 10−44 |
| GO:0016604 | Nuclear body | 63 | 742 | 5.02 × 10−26 | 47 | 742 | 8.81 × 10−16 |
| GO:0005694 | Chromosome | 60 | 950 | 1.13 × 10−18 | 60 | 950 | 2.48 × 10−20 |
| GO:0005681 | Spliceosomal complex | 53 | 187 | 1.13 × 10−44 | 33 | 187 | 6.24 × 10−23 |
| GO:0016607 | Nuclear speck | 47 | 381 | 2.12 × 10−25 | 32 | 381 | 9.22 × 10−14 |
| GO:0071013 | Catalytic step 2 spliceosome | 43 | 99 | 1.68 × 10−42 | 27 | 99 | 5.63 × 10−23 |
| GO:0000785 | Chromatin | 35 | 489 | 3.15 × 10−12 | 36 | 489 | 7.64 × 10−14 |
| GO:0000790 | Nuclear chromatin | 25 | 333 | 2.75 × 10−9 | 24 | 333 | 4.31 × 10−9 |
| GO:0000786 | Nucleosome | 17 | 106 | 7.01 × 10−11 | 15 | 106 | 2.51 × 10−9 |
| GO:0005635 | Nuclear envelope | 16 | 446 | 0.0079 | 15 | 446 | 0.0118 |
| GO:0000118 | Histone deacetylase complex | 8 | 60 | 6.32 × 10−5 | 7 | 60 | 0.00028 |
| GO:0016581 | NuRD complex | 6 | 14 | 3.05 × 10−6 | 5 | 14 | 4.20 × 10−5 |
| GO:0031105 | Septin complex | 4 | 6 | 6.96 × 10−5 | 4 | 6 | 5.79 × 10−5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barreiro-Alonso, A.; Lamas-Maceiras, M.; Lorenzo-Catoira, L.; Pardo, M.; Yu, L.; Choudhary, J.S.; Cerdán, M.E. HMGB1 Protein Interactions in Prostate and Ovary Cancer Models Reveal Links to RNA Processing and Ribosome Biogenesis through NuRD, THOC and Septin Complexes. Cancers 2021, 13, 4686. https://doi.org/10.3390/cancers13184686
Barreiro-Alonso A, Lamas-Maceiras M, Lorenzo-Catoira L, Pardo M, Yu L, Choudhary JS, Cerdán ME. HMGB1 Protein Interactions in Prostate and Ovary Cancer Models Reveal Links to RNA Processing and Ribosome Biogenesis through NuRD, THOC and Septin Complexes. Cancers. 2021; 13(18):4686. https://doi.org/10.3390/cancers13184686
Chicago/Turabian StyleBarreiro-Alonso, Aida, Mónica Lamas-Maceiras, Lidia Lorenzo-Catoira, Mercedes Pardo, Lu Yu, Jyoti S. Choudhary, and M. Esperanza Cerdán. 2021. "HMGB1 Protein Interactions in Prostate and Ovary Cancer Models Reveal Links to RNA Processing and Ribosome Biogenesis through NuRD, THOC and Septin Complexes" Cancers 13, no. 18: 4686. https://doi.org/10.3390/cancers13184686