DNA Methylation-Based Estimates of Circulating Leukocyte Composition for Predicting Colorectal Cancer Survival: A Prospective Cohort Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
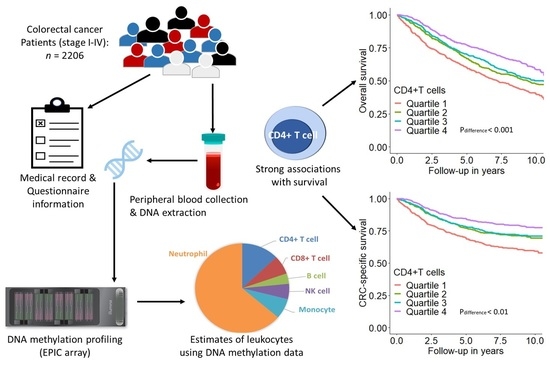
2.1. Study Design and Population
2.2. Data Collection
2.3. DNA Methylation and Leukocyte Composition Estimation
2.4. Statistical Methods
3. Results
3.1. Characteristics of the Study Population
3.2. Association of Leukocyte Composition with CRC Prognosis
3.3. Subgroup Analyses
3.4. Predictive Utility of CD4+ T Cell Proportion and All Leukocyte Proportions Combined
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Brenner, H.; Kloor, M.; Pox, C.P. Colorectal cancer. Lancet 2014, 383, 1490–1502. [Google Scholar] [CrossRef]
- Brenner, H.; Chen, C. The colorectal cancer epidemic: Challenges and opportunities for primary, secondary and tertiary prevention. Br. J. Cancer 2018, 119, 785–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galon, J.; Mlecnik, B.; Bindea, G.; Angell, H.K.; Berger, A.; Lagorce, C.; Lugli, A.; Zlobec, I.; Hartmann, A.; Bifulco, C.; et al. Towards the introduction of the ‘Immunoscore’ in the classification of malignant tumours. J. Pathol. 2014, 232, 199–209. [Google Scholar] [CrossRef] [Green Version]
- Gào, X.; Zhang, Y.; Boakye, D.; Li, X.; Chang-Claude, J.; Hoffmeister, M.; Brenner, H. Whole blood DNA methylation aging markers predict colorectal cancer survival: A prospective cohort study. Clin. Epigenetics 2020, 12, 184. [Google Scholar] [CrossRef]
- Kraus, S.; Arber, N. Inflammation and colorectal cancer. Curr. Opin. Pharmacol. 2009, 9, 405–410. [Google Scholar] [CrossRef]
- Terzić, J.; Grivennikov, S.; Karin, E.; Karin, M. Inflammation and colon cancer. Gastroenterology 2010, 138, 2101–2114.e2105. [Google Scholar] [CrossRef]
- Lança, T.; Silva-Santos, B. The split nature of tumor-infiltrating leukocytes: Implications for cancer surveillance and immunotherapy. Oncoimmunology 2012, 1, 717–725. [Google Scholar] [CrossRef] [Green Version]
- Zumsteg, A.; Christofori, G. Corrupt policemen: Inflammatory cells promote tumor angiogenesis. Curr. Opin. Oncol. 2009, 21, 60–70. [Google Scholar] [CrossRef]
- Li, M.X.; Liu, X.M.; Zhang, X.F.; Zhang, J.F.; Wang, W.L.; Zhu, Y.; Dong, J.; Cheng, J.W.; Liu, Z.W.; Ma, L.; et al. Prognostic role of neutrophil-to-lymphocyte ratio in colorectal cancer: A systematic review and meta-analysis. Int. J. Cancer 2014, 134, 2403–2413. [Google Scholar] [CrossRef]
- Ozdemir, Y.; Akin, M.L.; Sucullu, I.; Balta, A.Z.; Yucel, E. Pretreatment neutrophil/lymphocyte ratio as a prognostic aid in colorectal cancer. Asian Pac. J. Cancer Prev. 2014, 15, 2647–2650. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.Y.; Raghav, K.; Lieu, C.H.; Jiang, Z.Q.; Eng, C.; Vauthey, J.N.; Chang, G.J.; Qiao, W.; Morris, J.; Hong, D.; et al. Cytokine profile and prognostic significance of high neutrophil-lymphocyte ratio in colorectal cancer. Br. J. Cancer 2015, 112, 1088–1097. [Google Scholar] [CrossRef]
- Ikeguchi, M.; Ashida, K.; Saito, H. New Prognostic Indicator is Useful for Predicting the Survival of Patients with Unresectable Advanced Colorectal Cancer. Hepatogastroenterology 2015, 62, 859–862. [Google Scholar]
- Dimitriou, N.; Felekouras, E.; Karavokyros, I.; Alexandrou, A.; Pikoulis, E.; Griniatsos, J. Neutrophils to lymphocytes ratio as a useful prognosticator for stage II colorectal cancer patients. BMC Cancer 2018, 18, 1202. [Google Scholar] [CrossRef]
- Baron, U.; Türbachova, I.; Hellwag, A.; Eckhardt, F.; Berlin, K.; Hoffmuller, U.; Gardina, P.; Olek, S. DNA methylation analysis as a tool for cell typing. Epigenetics 2006, 1, 55–60. [Google Scholar] [CrossRef] [Green Version]
- Houseman, E.A.; Accomando, W.P.; Koestler, D.C.; Christensen, B.C.; Marsit, C.J.; Nelson, H.H.; Wiencke, J.K.; Kelsey, K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012, 13, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heiss, J.A.; Breitling, L.P.; Lehne, B.; Kooner, J.S.; Chambers, J.C.; Brenner, H. Training a model for estimating leukocyte composition using whole-blood DNA methylation and cell counts as reference. Epigenomics 2017, 9, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Salas, L.A.; Koestler, D.C.; Butler, R.A.; Hansen, H.M.; Wiencke, J.K.; Kelsey, K.T.; Christensen, B.C. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray. Genome Biol. 2018, 19, 64. [Google Scholar] [CrossRef]
- Brenner, H.; Chang-Claude, J.; Seiler, C.M.; Rickert, A.; Hoffmeister, M. Protection from colorectal cancer after colonoscopy: A population-based, case-control study. Ann. Intern. Med. 2011, 154, 22–30. [Google Scholar] [CrossRef]
- Brenner, H.; Chang-Claude, J.; Jansen, L.; Knebel, P.; Stock, C.; Hoffmeister, M. Reduced risk of colorectal cancer up to 10 years after screening, surveillance, or diagnostic colonoscopy. Gastroenterology 2014, 146, 709–717. [Google Scholar] [CrossRef] [Green Version]
- Hoffmeister, M.; Jansen, L.; Rudolph, A.; Toth, C.; Kloor, M.; Roth, W.; Bläker, H.; Chang-Claude, J.; Brenner, H. Statin use and survival after colorectal cancer: The importance of comprehensive confounder adjustment. J. Natl. Cancer Inst. 2015, 107, djv045. [Google Scholar] [CrossRef] [Green Version]
- Weigl, K.; Chang-Claude, J.; Knebel, P.; Hsu, L.; Hoffmeister, M.; Brenner, H. Strongly enhanced colorectal cancer risk stratification by combining family history and genetic risk score. Clin. Epidemiol. 2018, 10, 143–152. [Google Scholar] [CrossRef] [Green Version]
- Lehne, B.; Drong, A.W.; Loh, M.; Zhang, W.; Scott, W.R.; Tan, S.T.; Afzal, U.; Scott, J.; Jarvelin, M.R.; Elliott, P.; et al. A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol. 2015, 16, 37. [Google Scholar] [CrossRef] [Green Version]
- Salas, L.; Koestler, D. FlowSorted.Blood.EPIC: Illumina EPIC Data on Immunomagnetic Sorted Peripheral Adult Blood Cells. R Package Version 161. 2020. Available online: https://github.com/immunomethylomics/FlowSorted.Blood.EPIC (accessed on 1 April 2020).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: http://www.r-project.org/index.html (accessed on 1 April 2020).
- Borst, J.; Ahrends, T.; Bąbała, N.; Melief, C.J.M.; Kastenmüller, W. CD4+ T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2018, 18, 635–647. [Google Scholar] [CrossRef]
- Kuwahara, T.; Hazama, S.; Suzuki, N.; Yoshida, S.; Tomochika, S.; Nakagami, Y.; Matsui, H.; Shindo, Y.; Kanekiyo, S.; Tokumitsu, Y.; et al. Intratumoural-infiltrating CD4 + and FOXP3 + T cells as strong positive predictive markers for the prognosis of resectable colorectal cancer. Br. J. Cancer 2019, 121, 659–665. [Google Scholar] [CrossRef] [Green Version]
- Toor, S.M.; Murshed, K.; Al-Dhaheri, M.; Khawar, M.; Abu Nada, M.; Elkord, E. Immune Checkpoints in Circulating and Tumor-Infiltrating CD4+ T Cell Subsets in Colorectal Cancer Patients. Front. Immunol. 2019, 10, 2936. [Google Scholar] [CrossRef]
- Chaudhary, B.; Elkord, E. Regulatory T Cells in the Tumor Microenvironment and Cancer Progression: Role and Therapeutic Targeting. Vaccines 2016, 4, 28. [Google Scholar] [CrossRef] [Green Version]
- Betts, G.; Jones, E.; Junaid, S.; El-Shanawany, T.; Scurr, M.; Mizen, P.; Kumar, M.; Jones, S.; Rees, B.; Williams, G.; et al. Suppression of tumour-specific CD4+ T cells by regulatory T cells is associated with progression of human colorectal cancer. Gut 2012, 61, 1163–1171. [Google Scholar] [CrossRef] [Green Version]
- Reissfelder, C.; Stamova, S.; Gossmann, C.; Braun, M.; Bonertz, A.; Walliczek, U.; Grimm, M.; Rahbari, N.N.; Koch, M.; Saadati, M.; et al. Tumor-specific cytotoxic T lymphocyte activity determines colorectal cancer patient prognosis. J. Clin. Investig. 2015, 125, 739–751. [Google Scholar] [CrossRef]
- Glaire, M.A.; Domingo, E.; Sveen, A.; Bruun, J.; Nesbakken, A.; Nicholson, G.; Novelli, M.; Lawson, K.; Oukrif, D.; Kildal, W.; et al. Tumour-infiltrating CD8+ lymphocytes and colorectal cancer recurrence by tumour and nodal stage. Br. J. Cancer 2019, 121, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Yuen, G.J.; Demissie, E.; Pillai, S. B lymphocytes and cancer: A love-hate relationship. Trends Cancer 2016, 2, 747–757. [Google Scholar] [CrossRef] [Green Version]
- Berntsson, J.; Svensson, M.C.; Leandersson, K.; Nodin, B.; Micke, P.; Larsson, A.H.; Eberhard, J.; Jirström, K. The clinical impact of tumour-infiltrating lymphocytes in colorectal cancer differs by anatomical subsite: A cohort study. Int. J. Cancer 2017, 141, 1654–1666. [Google Scholar] [CrossRef]
- Edin, S.; Kaprio, T.; Hagström, J.; Larsson, P.; Mustonen, H.; Böckelman, C.; Strigård, K.; Gunnarsson, U.; Haglund, C.; Palmqvist, R. The Prognostic Importance of CD20+ B lymphocytes in Colorectal Cancer and the Relation to Other Immune Cell subsets. Sci. Rep. 2019, 9, 19997. [Google Scholar] [CrossRef]
- Tang, Y.P.; Xie, M.Z.; Li, K.Z.; Li, J.L.; Cai, Z.M.; Hu, B.L. Prognostic value of peripheral blood natural killer cells in colorectal cancer. BMC Gastroenterol. 2020, 20, 31. [Google Scholar] [CrossRef] [Green Version]
- Wen, S.; Chen, N.; Peng, J.; Ling, W.; Fang, Q.; Yin, S.F.; He, X.; Qiu, M.; Hu, Y. Peripheral monocyte counts predict the clinical outcome for patients with colorectal cancer: A systematic review and meta-analysis. Eur. J. Gastroenterol. Hepatol. 2019, 31, 1313–1321. [Google Scholar] [CrossRef]
- Grecian, R.; Whyte, M.K.B.; Walmsley, S.R. The role of neutrophils in cancer. Br. Med. Bull. 2018, 128, 5–14. [Google Scholar] [CrossRef] [Green Version]
- Mizuno, R.; Kawada, K.; Itatani, Y.; Ogawa, R.; Kiyasu, Y.; Sakai, Y. The Role of Tumor-Associated Neutrophils in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, H.L.; Chen, J.W.; Li, M.; Xiao, Y.B.; Fu, J.; Zeng, Y.X.; Cai, M.Y.; Xie, D. Increased intratumoral neutrophil in colorectal carcinomas correlates closely with malignant phenotype and predicts patients’ adverse prognosis. PLoS ONE 2012, 7, e30806. [Google Scholar] [CrossRef] [Green Version]
- Wikberg, M.L.; Ling, A.; Li, X.; Öberg, Å.; Edin, S.; Palmqvist, R. Neutrophil infiltration is a favorable prognostic factor in early stages of colon cancer. Hum. Pathol. 2017, 68, 193–202. [Google Scholar] [CrossRef]
- Dugué, P.A.; Bassett, J.K.; Joo, J.E.; Jung, C.H.; Ming Wong, E.; Moreno-Betancur, M.; Schmidt, D.; Makalic, E.; Li, S.; Severi, G.; et al. DNA methylation-based biological aging and cancer risk and survival: Pooled analysis of seven prospective studies. Int. J. Cancer 2018, 142, 1611–1619. [Google Scholar] [CrossRef]
- van Meir, H.; Nout, R.A.; Welters, M.J.; Loof, N.M.; de Kam, M.L.; van Ham, J.J.; Samuels, S.; Kenter, G.G.; Cohen, A.F.; Melief, C.J.; et al. Impact of (chemo)radiotherapy on immune cell composition and function in cervical cancer patients. Oncoimmunology 2017, 6, e1267095. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Kandimalla, R.; Huang, H.; Zhu, L.; Li, Y.; Gao, F.; Goel, A.; Wang, X. Molecular subtyping of colorectal cancer: Recent progress, new challenges and emerging opportunities. Semin. Cancer Biol. 2019, 55, 37–52. [Google Scholar] [CrossRef] [PubMed]




| Characteristics | n (%) * |
|---|---|
| Sex | |
| Women | 910 (41.2) |
| Men | 1296 (58.8) |
| Age at diagnosis | |
| [33, 65) years | 717 (32.5) |
| [65, 75) years | 776 (35.2) |
| [75, 96) years | 713 (32.3) |
| Tumor stage | |
| I | 400 (18.2) |
| II | 759 (34.6) |
| III | 726 (33.1) |
| IV | 309 (14.1) |
| Tumor subsite | |
| Distal colon † | 738 (33.5) |
| Proximal colon ‡ | 796 (36.1) |
| Rectum | 669 (30.4) |
| Body mass index at diagnosis | |
| <25 kg/m2 | 834 (38.0) |
| 25–30 kg/m2 | 932 (42.4) |
| >30 kg/m2 | 430 (19.6) |
| Smoking status | |
| Never | 907 (41.1) |
| Former | 947 (43.0) |
| Current | 350 (15.9) |
| Charlson comorbidity index | |
| 0 (no comorbidity) | 1281 (58.1) |
| 1 (mild comorbidity) | 479 (21.7) |
| 2+ (moderate comorbidity) | 446 (20.2) |
| Combination of Predictors | All-Cause Mortality | CRC-Specific Mortality | |
|---|---|---|---|
| All stages | Age + sex + stage | 0.739 (0.723, 0.754) | 0.809 (0.792, 0.825) |
| Age + sex + stage + CD4(+)T cell | 0.743 (0.728, 0.758) | 0.813 (0.797, 0.829) | |
| Age + sex + stage + all leukocyte subtypes | 0.744 (0.729, 0.759) | 0.814 (0.798, 0.830) | |
| Stage I and II | Age + sex | 0.693 (0.667, 0.718) | 0.612 (0.559, 0.666) |
| Age + sex + CD4(+)T cell | 0.703 (0.678, 0.729) | 0.630 (0.578, 0.682) | |
| Age + sex + all leukocyte subtypes | 0.706 (0.680, 0.731) | 0.643 (0.593, 0.693) | |
| Stage III | Age + sex | 0.653 (0.622, 0.683) | 0.608 (0.568, 0.648) |
| Age + sex + CD4(+)T cell | 0.659 (0.629, 0.689) | 0.622 (0.583, 0.660) | |
| Age + sex + all leukocyte subtypes | 0.662 (0.632, 0.691) | 0.626 (0.587, 0.665) | |
| Stage IV | Age + sex | 0.557 (0.520, 0.594) | 0.557 (0.519, 0.595) |
| Age + sex + CD4(+)T cell | 0.597 (0.562, 0.631) | 0.600 (0.564, 0.635) | |
| Age + sex + all leukocyte subtypes | 0.593 (0.557, 0.629) | 0.599 (0.562, 0.636) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gào, X.; Zhang, Y.; Li, X.; Jansen, L.; Alwers, E.; Bewerunge-Hudler, M.; Schick, M.; Chang-Claude, J.; Hoffmeister, M.; Brenner, H. DNA Methylation-Based Estimates of Circulating Leukocyte Composition for Predicting Colorectal Cancer Survival: A Prospective Cohort Study. Cancers 2021, 13, 2948. https://doi.org/10.3390/cancers13122948
Gào X, Zhang Y, Li X, Jansen L, Alwers E, Bewerunge-Hudler M, Schick M, Chang-Claude J, Hoffmeister M, Brenner H. DNA Methylation-Based Estimates of Circulating Leukocyte Composition for Predicting Colorectal Cancer Survival: A Prospective Cohort Study. Cancers. 2021; 13(12):2948. https://doi.org/10.3390/cancers13122948
Chicago/Turabian StyleGào, Xīn, Yan Zhang, Xiangwei Li, Lina Jansen, Elizabeth Alwers, Melanie Bewerunge-Hudler, Matthias Schick, Jenny Chang-Claude, Michael Hoffmeister, and Hermann Brenner. 2021. "DNA Methylation-Based Estimates of Circulating Leukocyte Composition for Predicting Colorectal Cancer Survival: A Prospective Cohort Study" Cancers 13, no. 12: 2948. https://doi.org/10.3390/cancers13122948