Identifying Cross-Scale Associations between Radiomic and Pathomic Signatures of Non-Small Cell Lung Cancer Subtypes: Preliminary Results
Abstract
:Simple Summary
Abstract
1. Introduction
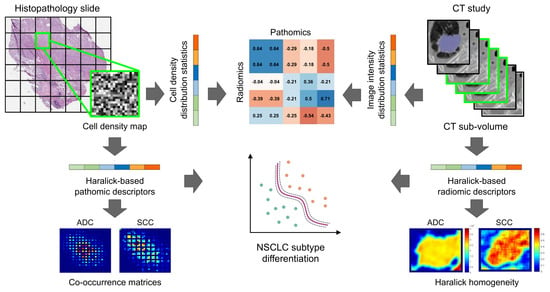
2. Materials and Methods
2.1. Digitized Pathology Data
2.2. Computed Tomography Imaging Data
2.3. Computing Cellular Density Map on Histopathology Slides
2.4. Pathomics Feature Extraction From Cellular Density Map
2.5. Radiomics Feature Extraction from CT Images
2.6. Experimental Design
2.6.1. Experiment 1: Identifying Pathomics Features to Differentiate ADC from SCC
2.6.2. Experiment 2: Identifying Radiomics Features to Differentiate ADC from SCC
2.6.3. Experiment 3: Exploratory Identification of Cross-Scale Pathomic-Radiomic Associations
2.7. Statistical Analysis
3. Results
3.1. Experiment 1: Identifying Pathomic Features to Differentiate ADC from SCC
3.2. Experiment 2: Identifying Radiomic Features to Differentiate ADC from SCC
3.3. Experiment 3: Exploratory Identification of Pathomic-Radiomic Associations for Differentiating ADC from SCC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| NSCLC | Non-small cell lung cancer |
| ADC | Adenocarcinoma |
| SCC | Squamous cell carcinoma |
| HV | Horizonal-vertical directions |
| D | Diagonal direction |
| SVM | Support vector machine |
| CT | Computer tomography |
| FDR | False discovery rate |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blandin Knight, S.; Crosbie, P.A.; Balata, H.; Chudziak, J.; Hussell, T.; Dive, C. Progress and prospects of early detection in lung cancer. Open Biol. 2017, 7, 170070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Travis, W.D.; Brambilla, E.; Noguchi, M.; Nicholson, A.G.; Geisinger, K.R.; Yatabe, Y.; Beer, D.G.; Powell, C.A.; Riely, G.J.; Van Schil, P.E.; et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J. Thorac. Oncol. 2011, 6, 244–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Li, M.; Huang, Y.; Ma, L.; Zhu, H.; Kong, L.; Yu, J. Clinical and radiological characteristics of central pulmonary adenocarcinoma: A comparison with central squamous cell carcinoma and small cell lung cancer and the impact on treatment response. OncoTargets Ther. 2018, 11, 2509. [Google Scholar] [CrossRef] [Green Version]
- Zappa, C.; Mousa, S.A. Non-small cell lung cancer: Current treatment and future advances. Transl. Lung Cancer Res. 2016, 5, 288–300. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Feng, W.; Wu, Z.; Liu, M.; Zhang, F.; Liang, Z.; Cui, C.; Huang, J.; Li, X.; Guo, X. Intra-tumoural heterogeneity characterization through texture and colour analysis for differentiation of non-small cell lung carcinoma subtypes. Phys. Med. Biol. 2018, 63, 165018. [Google Scholar] [CrossRef]
- Asamura, H.; Goya, T.; Koshiishi, Y.; Sohara, Y.; Eguchi, K.; Mori, M.; Nakanishi, Y.; Tsuchiya, R.; Shimokata, K.; Inoue, H.; et al. A Japanese Lung Cancer Registry study: Prognosis of 13,010 resected lung cancers. J. Thorac. Oncol. 2008, 3, 46–52. [Google Scholar] [CrossRef] [Green Version]
- Reck, M.; Rabe, K.F. Precision diagnosis and treatment for advanced non–small-cell lung cancer. N. Engl. J. Med. 2017, 377, 849–861. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Parmar, C.; Grossmann, P.; Quackenbush, J.; Lambin, P.; Bussink, J.; Mak, R.; Aerts, H.J. Exploratory study to identify radiomics classifiers for lung cancer histology. Front. Oncol. 2016, 6, 71. [Google Scholar] [CrossRef] [Green Version]
- Lambin, P.; Leijenaar, R.T.; Deist, T.M.; Peerlings, J.; De Jong, E.E.; Van Timmeren, J.; Sanduleanu, S.; Larue, R.T.; Even, A.J.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gnep, K.; Fargeas, A.; Gutiérrez-Carvajal, R.E.; Commandeur, F.; Mathieu, R.; Ospina, J.D.; Rolland, Y.; Rohou, T.; Vincendeau, S.; Hatt, M.; et al. Haralick textural features on T2-weighted MRI are associated with biochemical recurrence following radiotherapy for peripheral zone prostate cancer. J. Magn. Reson. Imaging 2017, 45, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Wibmer, A.; Hricak, H.; Gondo, T.; Matsumoto, K.; Veeraraghavan, H.; Fehr, D.; Zheng, J.; Goldman, D.; Moskowitz, C.; Fine, S.W.; et al. Haralick texture analysis of prostate MRI: Utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur. Radiol. 2015, 25, 2840–2850. [Google Scholar] [CrossRef]
- Khalvati, F.; Wong, A.; Haider, M.A. Automated prostate cancer detection via comprehensive multi-parametric magnetic resonance imaging texture feature models. BMC Med. Imaging 2015, 15, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Zhu, Y.; Burnside, E.S.; Drukker, K.; Hoadley, K.A.; Fan, C.; Conzen, S.D.; Whitman, G.J.; Sutton, E.J.; Net, J.M.; et al. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of MammaPrint, Oncotype DX, and PAM50 gene assays. Radiology 2016, 281, 382–391. [Google Scholar] [CrossRef] [Green Version]
- Rahbar, H.; McDonald, E.S.; Lee, J.M.; Partridge, S.C.; Lee, C.I. How can advanced imaging be used to mitigate potential breast cancer overdiagnosis? Acad. Radiol. 2016, 23, 768–773. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Kato, F.; Oyama-Manabe, N.; Li, R.; Cui, Y.; Tha, K.K.; Yamashita, H.; Kudo, K.; Shirato, H. Identifying triple-negative breast cancer using background parenchymal enhancement heterogeneity on dynamic contrast-enhanced MRI: A pilot radiomics study. PLoS ONE 2015, 10, e0143308. [Google Scholar] [CrossRef]
- Coroller, T.P.; Grossmann, P.; Hou, Y.; Velazquez, E.R.; Leijenaar, R.T.; Hermann, G.; Lambin, P.; Haibe-Kains, B.; Mak, R.H.; Aerts, H.J. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother. Oncol. 2015, 114, 345–350. [Google Scholar] [CrossRef]
- Depeursinge, A.; Yanagawa, M.; Leung, A.N.; Rubin, D.L. Predicting adenocarcinoma recurrence using computational texture models of nodule components in lung CT. Med. Phys. 2015, 42, 2054–2063. [Google Scholar] [CrossRef] [Green Version]
- Thawani, R.; McLane, M.; Beig, N.; Ghose, S.; Prasanna, P.; Velcheti, V.; Madabhushi, A. Radiomics and radiogenomics in lung cancer: A review for the clinician. Lung Cancer 2018, 115, 34–41. [Google Scholar] [CrossRef]
- Kickingereder, P.; Burth, S.; Wick, A.; Götz, M.; Eidel, O.; Schlemmer, H.P.; Maier-Hein, K.H.; Wick, W.; Bendszus, M.; Radbruch, A.; et al. Radiomic profiling of glioblastoma: Identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology 2016, 280, 880–889. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Prasanna, P.; Wolansky, L.; Pinho, M.; Cohen, M.; Nayate, A.; Gupta, A.; Singh, G.; Hatanpaa, K.; Sloan, A.; et al. Computer-extracted texture features to distinguish cerebral radionecrosis from recurrent brain tumors on multiparametric MRI: A feasibility study. Am. J. Neuroradiol. 2016, 37, 2231–2236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antunes, J.T.; Ofshteyn, A.; Bera, K.; Wang, E.Y.; Brady, J.T.; Willis, J.E.; Friedman, K.A.; Marderstein, E.L.; Kalady, M.F.; Stein, S.L.; et al. Radiomic Features of Primary Rectal Cancers on Baseline T2-Weighted MRI Are Associated With Pathologic Complete Response to Neoadjuvant Chemoradiation: A Multisite Study. J. Magn. Reson. Imaging 2020. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Huang, Y.; He, L.; Chen, X.; Ma, Z.; Dong, D.; Tian, J.; Liang, C.; Liu, Z. The development and validation of a CT-based radiomics signature for the preoperative discrimination of stage I-II and stage III-IV colorectal cancer. Oncotarget 2016, 7, 31401. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.Q.; Liang, C.H.; He, L.; Tian, J.; Liang, C.S.; Chen, X.; Ma, Z.L.; Liu, Z.Y. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J. Clin. Oncol. 2016, 34, 2157–2164. [Google Scholar] [CrossRef]
- Antunes, J.; Viswanath, S.; Rusu, M.; Valls, L.; Hoimes, C.; Avril, N.; Madabhushi, A. Radiomics analysis on FLT-PET/MRI for characterization of early treatment response in renal cell carcinoma: A proof-of-concept study. Transl. Oncol. 2016, 9, 155–162. [Google Scholar] [CrossRef] [Green Version]
- Parmar, C.; Grossmann, P.; Rietveld, D.; Rietbergen, M.M.; Lambin, P.; Aerts, H.J. Radiomic machine-learning classifiers for prognostic biomarkers of head and neck cancer. Front. Oncol. 2015, 5, 272. [Google Scholar] [CrossRef] [Green Version]
- Parmar, C.; Leijenaar, R.T.; Grossmann, P.; Velazquez, E.R.; Bussink, J.; Rietveld, D.; Rietbergen, M.M.; Haibe-Kains, B.; Lambin, P.; Aerts, H.J. Radiomic feature clusters and prognostic signatures specific for lung and head & neck cancer. Sci. Rep. 2015, 5, 11044. [Google Scholar]
- Haga, A.; Takahashi, W.; Aoki, S.; Nawa, K.; Yamashita, H.; Abe, O.; Nakagawa, K. Classification of early stage non-small cell lung cancers on computed tomographic images into histological types using radiomic features: Interobserver delineation variability analysis. Radiol. Phys. Technol. 2018, 11, 27–35. [Google Scholar] [CrossRef]
- Bashir, U.; Kawa, B.; Siddique, M.; Mak, S.M.; Nair, A.; Mclean, E.; Bille, A.; Goh, V.; Cook, G. Non-invasive classification of non-small cell lung cancer: A comparison between random forest models utilising radiomic and semantic features. Br. J. Radiol. 2019, 92, 20190159. [Google Scholar] [CrossRef]
- Linning, E.; Lu, L.; Li, L.; Yang, H.; Schwartz, L.H.; Zhao, B. Radiomics for classifying histological subtypes of lung cancer based on multiphasic contrast-enhanced computed tomography. J. Comput. Assist. Tomogr. 2019, 43, 300–306. [Google Scholar]
- Zhu, X.; Dong, D.; Chen, Z.; Fang, M.; Zhang, L.; Song, J.; Yu, D.; Zang, Y.; Liu, Z.; Shi, J.; et al. Radiomic signature as a diagnostic factor for histologic subtype classification of non-small cell lung cancer. Eur. Radiol. 2018, 28, 2772–2778. [Google Scholar] [CrossRef] [PubMed]
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial intelligence in digital pathology—New tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Kurc, T.; Sharma, A.; Almeida, J.S.; Saltz, J. The emergence of pathomics. Curr. Pathobiol. Rep. 2019, 7, 73–84. [Google Scholar] [CrossRef] [Green Version]
- Tran, W.T.; Jerzak, K.; Lu, F.I.; Klein, J.; Tabbarah, S.; Lagree, A.; Wu, T.; Rosado-Mendez, I.; Law, E.; Saednia, K.; et al. Personalized breast cancer treatments using artificial intelligence in radiomics and pathomics. J. Med. Imaging Radiat. Sci. 2019, 50, S32–S41. [Google Scholar] [CrossRef] [Green Version]
- Saltz, J.; Almeida, J.; Gao, Y.; Sharma, A.; Bremer, E.; DiPrima, T.; Saltz, M.; Kalpathy-Cramer, J.; Kurc, T. Towards generation, management, and exploration of combined radiomics and pathomics datasets for cancer research. AMIA Summits Transl. Sci. Proc. 2017, 2017, 85. [Google Scholar]
- Corredor, G.; Wang, X.; Zhou, Y.; Lu, C.; Fu, P.; Syrigos, K.; Rimm, D.L.; Yang, M.; Romero, E.; Schalper, K.A.; et al. Spatial architecture and arrangement of tumor-infiltrating lymphocytes for predicting likelihood of recurrence in early-stage non–small cell lung cancer. Clin. Cancer Res. 2019, 25, 1526–1534. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Janowczyk, A.; Zhou, Y.; Thawani, R.; Fu, P.; Schalper, K.; Velcheti, V.; Madabhushi, A. Prediction of recurrence in early stage non-small cell lung cancer using computer extracted nuclear features from digital H&E images. Sci. Rep. 2017, 7, 1–10. [Google Scholar]
- Yao, J.; Ganti, D.; Luo, X.; Xiao, G.; Xie, Y.; Yan, S.; Huang, J. Computer-assisted diagnosis of lung cancer using quantitative topology features. In International Workshop on Machine Learning in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2015; pp. 288–295. [Google Scholar]
- Wang, C.W.; Yu, C.P. Automated morphological classification of lung cancer subtypes using H&E tissue images. Mach. Vis. Appl. 2013, 24, 1383–1391. [Google Scholar]
- Khosravi, P.; Kazemi, E.; Imielinski, M.; Elemento, O.; Hajirasouliha, I. Deep convolutional neural networks enable discrimination of heterogeneous digital pathology images. EBioMedicine 2018, 27, 317–328. [Google Scholar] [CrossRef] [Green Version]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559. [Google Scholar] [CrossRef] [PubMed]
- Penzias, G.; Singanamalli, A.; Elliott, R.; Gollamudi, J.; Shih, N.; Feldman, M.; Stricker, P.D.; Delprado, W.; Tiwari, S.; Böhm, M.; et al. Identifying the morphologic basis for radiomic features in distinguishing different Gleason grades of prostate cancer on MRI: Preliminary findings. PLoS ONE 2018, 13, e0200730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, A.; Watson, G.; Myint, E.; Sved, P.; McEntee, M.; Bourne, R. Changes in epithelium, stroma, and lumen space correlate more strongly with Gleason pattern and are stronger predictors of prostate ADC changes than cellularity metrics. Radiology 2015, 277, 751–762. [Google Scholar] [CrossRef]
- Bobholz, S.A.; Lowman, A.K.; Barrington, A.; Brehler, M.; McGarry, S.; Cochran, E.J.; Connelly, J.; Mueller, W.M.; Agarwal, M.; O’Neill, D.; et al. Radiomic Features of Multiparametric MRI Present Stable Associations With Analogous Histological Features in Patients With Brain Cancer. Tomography 2020, 6, 160. [Google Scholar] [PubMed]
- Braman, N.; Prasanna, P.; Whitney, J.; Singh, S.; Beig, N.; Etesami, M.; Bates, D.D.; Gallagher, K.; Bloch, B.N.; Vulchi, M.; et al. Association of peritumoral radiomics with tumor biology and pathologic response to preoperative targeted therapy for HER2 (ERBB2)–positive breast cancer. JAMA Netw. Open 2019, 2, e192561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lederlin, M.; Puderbach, M.; Muley, T.; Schnabel, P.A.; Stenzinger, A.; Kauczor, H.U.; Heussel, C.P.; Herth, F.J.; Hoffmann, H.; Dienemann, H.; et al. Correlation of radio-and histomorphological pattern of pulmonary adenocarcinoma. Eur. Respir. J. 2013, 41, 943–951. [Google Scholar] [CrossRef] [Green Version]
- Khorrami, M.; Prasanna, P.; Gupta, A.; Patil, P.; Velu, P.D.; Thawani, R.; Corredor, G.; Alilou, M.; Bera, K.; Fu, P.; et al. Changes in CT Radiomic Features Associated with Lymphocyte Distribution Predict Overall Survival and Response to Immunotherapy in Non–Small Cell Lung Cancer. Cancer Immunol. Res. 2020, 8, 108–119. [Google Scholar] [CrossRef]
- Solis, L.M.; Behrens, C.; Raso, M.G.; Lin, H.Y.; Kadara, H.; Yuan, P.; Galindo, H.; Tang, X.; Lee, J.J.; Kalhor, N.; et al. Histologic patterns and molecular characteristics of lung adenocarcinoma associated with clinical outcome. Cancer 2012, 118, 2889–2899. [Google Scholar] [CrossRef] [Green Version]
- Santos, M.K.; Muley, T.; Warth, A.; de Paula, W.D.; Lederlin, M.; Schnabel, P.A.; Schlemmer, H.P.; Kauczor, H.U.; Heussel, C.P.; Puderbach, M. Morphological computed tomography features of surgically resectable pulmonary squamous cell carcinomas: Impact on prognosis and comparison with adenocarcinomas. Eur. J. Radiol. 2014, 83, 1275–1281. [Google Scholar] [CrossRef]
- Hollings, N.; Shaw, P. Diagnostic imaging of lung cancer. Eur. Respir. J. 2002, 19, 722–742. [Google Scholar] [CrossRef] [Green Version]
- National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). Radiology Data from the Clinical Proteomic Tumor Analysis Consortium Lung Squamous Cell Carcinoma [CPTAC-LSCC] Collection; National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC): Fayetteville, AR, USA, 2018. [CrossRef]
- National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). Radiology Data from the Clinical Proteomic Tumor Analysis Consortium Lung Adenocarcinoma [CPTAC-LUAD] Collection; National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC): Fayetteville, AR, USA, 2018. [CrossRef]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M.; Cancer Genome Atlas Research Network; et al. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013, 45, 1113. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J.W.L.; Rios Velazquez, E.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Data From NSCLC-Radiomics-Genomics; Cancer Imaging Archive: Fayetteville, AR, USA, 2015. [Google Scholar] [CrossRef]
- Veta, M.; Van Diest, P.J.; Kornegoor, R.; Huisman, A.; Viergever, M.A.; Pluim, J.P. Automatic nuclei segmentation in H&E stained breast cancer histopathology images. PLoS ONE 2013, 8, e70221. [Google Scholar]
- Infante, M.; Berghmans, T.; Heuvelmans, M.A.; Hillerdal, G.; Oudkerk, M. Slow-growing lung cancer as an emerging entity: From screening to clinical management. Eur. Respir. J. 2013, 42, 1706–1722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haralick, R.M. Statistical and structural approaches to texture. Proc. IEEE 1979, 67, 786–804. [Google Scholar] [CrossRef]
- Corder, G.W.; Foreman, D.I. Nonparametric Statistics: A Step-by-Step Approach; John Wiley & Sons: Hoboken, NJ, USA, 2014. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ganeshan, B.; Goh, V.; Mandeville, H.C.; Ng, Q.S.; Hoskin, P.J.; Miles, K.A. Non–small cell lung cancer: Histopathologic correlates for texture parameters at CT. Radiology 2013, 266, 326–336. [Google Scholar] [CrossRef]
- Snoeckx, A.; Reyntiens, P.; Pauwels, P.; Van Schil, P.; Spinhoven, M.; Parizel, P. Radiologic-Pathologic Correlation in Lung Cancer Presenting as a Subsolid Nodule: Room for Improvement? Clin. Oncol. 2017, 2, 1290. [Google Scholar]









| Parameter | CPTAC (117) | TCGA (54) | |
|---|---|---|---|
| Magnification | 20× | 117 | 29 |
| 40× | 0 | 25 | |
| Resolution (μm∖px) | 0.252 | 0 | 25 |
| 0.494 | 117 | 0 | |
| 0.502 | 0 | 29 | |
| Gender | Male | 82 | 23 |
| Female | 35 | 31 | |
| Age at diagnosis | 65.1 ± 9.3 | 68.6 ± 10.00 | |
| Grade | Well differentiated | 8 | NA |
| Moderately differentiated | 109 | NA | |
| Poorly differentiated | 0 | NA | |
| Undifferentiated | 0 | NA | |
| Pathologic Stage | I | 58 | 19 |
| II | 38 | 18 | |
| III | 21 | 12 | |
| IV | 0 | 3 | |
| Discrepancy | 3 | 2 | |
| NSCLC subtype | ADC | 61 | 25 |
| SCC | 56 | 29 | |
| Parameter | NSCLC-R (101) | TCGA (45) | |
|---|---|---|---|
| In-plane Resolution (mm) | 0.97 | 0.6–0.97 | |
| Slice Thickness (mm) | 3 | 2–5 | |
| Field of view (px) | 512 × 512 | 357–512 × 357–512 | |
| Scanner | CMS, Inc. | 11 | 0 |
| Siemens | 90 | 10 | |
| GE Medical Systems | 0 | 33 | |
| Philips | 0 | 2 | |
| Patient Position | Head First Supine (HFS) | NA | |
| Gender | Male | 64 | 20 |
| Female | 37 | 25 | |
| Age at diagnosis | 68.5 ± 10.4 | 69.3 ± 9.8 | |
| Pathologic Stage | I | 17 | 16 |
| II | 13 | 15 | |
| III | 71 | 9 | |
| IV | 0 | 3 | |
| Discrepancy | 0 | 2 | |
| Subtype | ADC | 49 | 19 |
| SCC | 52 | 26 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alvarez-Jimenez, C.; Sandino, A.A.; Prasanna, P.; Gupta, A.; Viswanath, S.E.; Romero, E. Identifying Cross-Scale Associations between Radiomic and Pathomic Signatures of Non-Small Cell Lung Cancer Subtypes: Preliminary Results. Cancers 2020, 12, 3663. https://doi.org/10.3390/cancers12123663
Alvarez-Jimenez C, Sandino AA, Prasanna P, Gupta A, Viswanath SE, Romero E. Identifying Cross-Scale Associations between Radiomic and Pathomic Signatures of Non-Small Cell Lung Cancer Subtypes: Preliminary Results. Cancers. 2020; 12(12):3663. https://doi.org/10.3390/cancers12123663
Chicago/Turabian StyleAlvarez-Jimenez, Charlems, Alvaro A. Sandino, Prateek Prasanna, Amit Gupta, Satish E. Viswanath, and Eduardo Romero. 2020. "Identifying Cross-Scale Associations between Radiomic and Pathomic Signatures of Non-Small Cell Lung Cancer Subtypes: Preliminary Results" Cancers 12, no. 12: 3663. https://doi.org/10.3390/cancers12123663