Pathways, Processes, and Candidate Drugs Associated with a Hoxa Cluster-Dependency Model of Leukemia
Abstract
:1. Introduction
2. Results
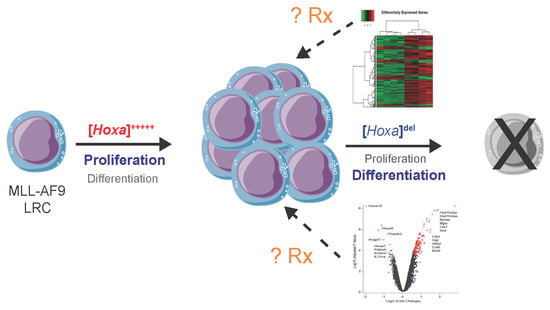
2.1. Establishment and Validation of Conditional Hoxa Leukemia Models
2.2. Reduced Leukemia Colony Formation Following IFNα-Induced Hoxa Deletion
2.3. Extension in Survival Following In Vivo Deletion of Hoxa Cluster
2.4. Hoxadel Signature and Associated Pathways
2.5. Gene Set Enrichment Analysis
3. Discussion
4. Materials and Methods
4.1. Mouse Strains
4.2. Generation of Primary Leukemias and Cell Lines
4.3. Deletion of the Hoxa Cluster
4.4. Gene Expression Analysis and Bioinformatics
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chessells, J.M.; Harrison, C.J.; Kempski, H.; Webb, D.K.; Wheatley, K.; Hann, I.M.; Stevens, R.F.; Harrison, G.; Gibson, B.E. Clinical features, cytogenetics and outcome in acute lymphoblastic and myeloid leukaemia of infancy: Report from the MRC Childhood Leukaemia working party. Leukemia 2002, 16, 776–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muntean, A.G. Mechanisms of mixed-lineage leukemia. Int. J. Hematol. Oncol. 2013, 2, 207–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubnitz, J.E.; Link, M.P.; Shuster, J.J.; Carroll, A.J.; Hakami, N.; Frankel, L.S.; Pullen, D.J.; Cleary, M.L. Frequency and prognostic significance of HRX rearrangements in infant acute lymphoblastic leukemia: A Pediatric Oncology Group study. Blood 1994, 84, 570–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tien, H.F.; Hsiao, C.H.; Tang, J.L.; Tsay, W.; Hu, C.H.; Kuo, Y.Y.; Wang, C.H.; Chen, Y.C.; Shen, M.C.; Lin, D.T.; et al. Characterization of acute myeloid leukemia with MLL rearrangements--no increase in the incidence of coexpression of lymphoid-associated antigens on leukemic blasts. Leukemia 2000, 14, 1025–1030. [Google Scholar] [CrossRef] [Green Version]
- Felix, C.A. Leukemias related to treatment with DNA topoisomerase II inhibitors. Med. Pediatric Oncol. 2001, 36, 525–535. [Google Scholar] [CrossRef]
- Pui, C.H.; Relling, M.V. Topoisomerase II inhibitor-related acute myeloid leukaemia. Br. J. Haematol. 2000, 109, 13–23. [Google Scholar] [CrossRef]
- Schoch, C.; Schnittger, S.; Klaus, M.; Kern, W.; Hiddemann, W.; Haferlach, T. AML with 11q23/MLL abnormalities as defined by the WHO classification: Incidence, partner chromosomes, FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood 2003, 102, 2395–2402. [Google Scholar] [CrossRef] [Green Version]
- Super, H.J.; McCabe, N.R.; Thirman, M.J.; Larson, R.A.; Le Beau, M.M.; Pedersen-Bjergaard, J.; Philip, P.; Diaz, M.O.; Rowley, J.D. Rearrangements of the MLL gene in therapy-related acute myeloid leukemia in patients previously treated with agents targeting DNA-topoisomerase II. Blood 1993, 82, 3705–3711. [Google Scholar] [CrossRef] [Green Version]
- Breen, T.R.; Harte, P.J. Molecular characterization of the trithorax gene, a positive regulator of homeotic gene expression in Drosophila. Mech. Dev. 1991, 35, 113–127. [Google Scholar] [CrossRef]
- Breen, T.R.; Harte, P.J. Trithorax regulates multiple homeotic genes in the bithorax and Antennapedia complexes and exerts different tissue-specific, parasegment-specific and promoter-specific effects on each. Development 1993, 117, 119–134. [Google Scholar]
- Duboule, D.; Dolle, P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 1989, 8, 1497–1505. [Google Scholar] [CrossRef] [PubMed]
- Graham, A.; Papalopulu, N.; Krumlauf, R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell 1989, 57, 367–378. [Google Scholar] [CrossRef]
- Yu, B.D.; Hanson, R.D.; Hess, J.L.; Horning, S.E.; Korsmeyer, S.J. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc. Natl. Acad. Sci. USA 1998, 95, 10632–10636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, L.H.; Slany, R.; Cui, X.; Cleary, M.L.; Mason, D.Y. The HRX proto-oncogene product is widely expressed in human tissues and localizes to nuclear structures. Blood 1997, 89, 3361–3370. [Google Scholar] [CrossRef] [Green Version]
- Ernst, P.; Mabon, M.; Davidson, A.J.; Zon, L.I.; Korsmeyer, S.J. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr. Biol. 2004, 14, 2063–2069. [Google Scholar] [CrossRef] [Green Version]
- Kawagoe, H.; Humphries, R.K.; Blair, A.; Sutherland, H.J.; Hogge, D.E. Expression of HOX genes, HOX cofactors, and MLL in phenotypically and functionally defined subpopulations of leukemic and normal human hematopoietic cells. Leukemia 1999, 13, 687–698. [Google Scholar] [CrossRef] [Green Version]
- Yu, B.D.; Hess, J.L.; Horning, S.E.; Brown, G.A.; Korsmeyer, S.J. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 1995, 378, 505–508. [Google Scholar] [CrossRef]
- Jude, C.D.; Climer, L.; Xu, D.; Artinger, E.; Fisher, J.K.; Ernst, P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell 2007, 1, 324–337. [Google Scholar] [CrossRef] [Green Version]
- McMahon, K.A.; Hiew, S.Y.; Hadjur, S.; Veiga-Fernandes, H.; Menzel, U.; Price, A.J.; Kioussis, D.; Williams, O.; Brady, H.J. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell 2007, 1, 338–345. [Google Scholar] [CrossRef] [Green Version]
- Abramovich, C.; Humphries, R.K. Hox regulation of normal and leukemic hematopoietic stem cells. Curr. Opin. Hematol. 2005, 12, 210–216. [Google Scholar] [CrossRef]
- Armstrong, S.A.; Staunton, J.E.; Silverman, L.B.; Pieters, R.; den Boer, M.L.; Minden, M.D.; Sallan, S.E.; Lander, E.S.; Golub, T.R.; Korsmeyer, S.J. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet. 2002, 30, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Dickson, G.J.; Liberante, F.G.; Kettyle, L.M.; O’Hagan, K.A.; Finnegan, D.P.; Bullinger, L.; Geerts, D.; McMullin, M.F.; Lappin, T.R.; Mills, K.I.; et al. HOXA/PBX3 knockdown impairs growth and sensitizes cytogenetically normal acute myeloid leukemia cells to chemotherapy. Haematologica 2013, 98, 1216–1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrando, A.A.; Armstrong, S.A.; Neuberg, D.S.; Sallan, S.E.; Silverman, L.B.; Korsmeyer, S.J.; Look, A.T. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: Dominance of HOX dysregulation. Blood 2003, 102, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Haferlach, C.; Haferlach, T. Another piece of the AML puzzle. Blood 2013, 122, 2532–2534. [Google Scholar] [CrossRef] [Green Version]
- Milne, T.A.; Martin, M.E.; Brock, H.W.; Slany, R.K.; Hess, J.L. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005, 65, 11367–11374. [Google Scholar] [CrossRef] [Green Version]
- Ayton, P.M.; Cleary, M.L. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003, 17, 2298–2307. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.R.; Hudson, W.A.; Chen, W.; Nishiuchi, R.; Yao, Q.; Kersey, J.H. Hoxa9 influences the phenotype but not the incidence of Mll-AF9 fusion gene leukemia. Blood 2004, 103, 1823–1828. [Google Scholar] [CrossRef] [Green Version]
- So, C.W.; Karsunky, H.; Wong, P.; Weissman, I.L.; Cleary, M.L. Leukemic transformation of hematopoietic progenitors by MLL-GAS7 in the absence of Hoxa7 or Hoxa9. Blood 2004, 103, 3192–3199. [Google Scholar] [CrossRef]
- Kmita, M.; Tarchini, B.; Zakany, J.; Logan, M.; Tabin, C.J.; Duboule, D. Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature 2005, 435, 1113–1116. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.N.; Wang, Z.C.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bower, H.; Bjorkholm, M.; Dickman, P.W.; Hoglund, M.; Lambert, P.C.; Andersson, T.M. Life Expectancy of Patients With Chronic Myeloid Leukemia Approaches the Life Expectancy of the General Population. J. Clin. Oncol. 2016, 34, 2851–2857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grier, D.G.; Thompson, A.; Kwasniewska, A.; McGonigle, G.J.; Halliday, H.L.; Lappin, T.R. The pathophysiology of HOX genes and their role in cancer. J. Pathol. 2005, 205, 154–171. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, R.A.; Pettengell, R.; Pandha, H.S.; Morgan, R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia 2013, 27, 1000–1008. [Google Scholar] [CrossRef] [Green Version]
- Crooks, G.M.; Fuller, J.; Petersen, D.; Izadi, P.; Malik, P.; Pattengale, P.K.; Kohn, D.B.; Gasson, J.C. Constitutive HOXA5 expression inhibits erythropoiesis and increases myelopoiesis from human hematopoietic progenitors. Blood 1999, 94, 519–528. [Google Scholar] [CrossRef]
- Fuller, J.F.; McAdara, J.; Yaron, Y.; Sakaguchi, M.; Fraser, J.K.; Gasson, J.C. Characterization of HOX gene expression during myelopoiesis: Role of HOX A5 in lineage commitment and maturation. Blood 1999, 93, 3391–3400. [Google Scholar] [CrossRef]
- Dickson, G.J.; Kwasniewska, A.; Mills, K.I.; Lappin, T.R.; Thompson, A. Hoxa6 potentiates short-term hemopoietic cell proliferation and extended self-renewal. Exp. Hematol. 2009, 37, 322–333. [Google Scholar] [CrossRef]
- Magnusson, M.; Brun, A.C.; Miyake, N.; Larsson, J.; Ehinger, M.; Bjornsson, J.M.; Wutz, A.; Sigvardsson, M.; Karlsson, S. HOXA10 is a critical regulator for hematopoietic stem cells and erythroid/megakaryocyte development. Blood 2007, 109, 3687–3696. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, H.J.; Helgason, C.D.; Sauvageau, G.; Fong, S.; Izon, D.J.; Humphries, R.K.; Largman, C. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood 1997, 89, 1922–1930. [Google Scholar] [CrossRef]
- Magnusson, M.; Brun, A.C.; Lawrence, H.J.; Karlsson, S. Hoxa9/hoxb3/hoxb4 compound null mice display severe hematopoietic defects. Exp. Hematol. 2007, 35, 1421–1428. [Google Scholar] [CrossRef]
- Thorsteinsdottir, U.; Mamo, A.; Kroon, E.; Jerome, L.; Bijl, J.; Lawrence, H.J.; Humphries, K.; Sauvageau, G. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood 2002, 99, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Faber, J.; Krivtsov, A.V.; Stubbs, M.C.; Wright, R.; Davis, T.N.; van den Heuvel-Eibrink, M.; Zwaan, C.M.; Kung, A.L.; Armstrong, S.A. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood 2009, 113, 2375–2385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horton, S.J.; Grier, D.G.; McGonigle, G.J.; Thompson, A.; Morrow, M.; De Silva, I.; Moulding, D.A.; Kioussis, D.; Lappin, T.R.; Brady, H.J.; et al. Continuous MLL-ENL expression is necessary to establish a “Hox Code” and maintain immortalization of hematopoietic progenitor cells. Cancer Res. 2005, 65, 9245–9252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, N.; Yang, L.Y.; Xu, Z.P. Long non-coding RNA HOTTIP is able to predict poor prognosis in various neoplasms: A meta-analysis. Mol. Clin. Oncol. 2017, 7, 263–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Y.; Wang, J.; Wu, F.; Song, Y.; Zhao, S.; Zhang, Q. Long non-coding RNA HOXA-AS2 promotes proliferation and invasion of breast cancer by acting as a miR-520c-3p sponge. Oncotarget 2017, 8, 46090–46103. [Google Scholar] [CrossRef] [PubMed]
- Lebert-Ghali, C.E.; Fournier, M.; Kettyle, L.; Thompson, A.; Sauvageau, G.; Bijl, J.J. Hoxa cluster genes determine the proliferative activity of adult mouse hematopoietic stem and progenitor cells. Blood 2016, 127, 87–90. [Google Scholar] [CrossRef] [Green Version]
- Maraka, S.; Groves, M.D.; Mammoser, A.G.; Melguizo-Gavilanes, I.; Conrad, C.A.; Tremont-Lukats, I.W.; Loghin, M.E.; O’Brien, B.J.; Puduvalli, V.K.; Sulman, E.P.; et al. Phase 1 lead-in to a phase 2 factorial study of temozolomide plus memantine, mefloquine, and metformin as postradiation adjuvant therapy for newly diagnosed glioblastoma. Cancer 2019, 125, 424–433. [Google Scholar] [CrossRef] [Green Version]
- Nolte, E.; Wach, S.; Silva, I.T.; Lukat, S.; Ekici, A.B.; Munkert, J.; Muller-Uri, F.; Kreis, W.; Oliveira Simoes, C.M.; Vera, J.; et al. A new semisynthetic cardenolide analog 3beta-[2-(1-amantadine)- 1-on-ethylamine]-digitoxigenin (AMANTADIG) affects G2/M cell cycle arrest and miRNA expression profiles and enhances proapoptotic survivin-2B expression in renal cell carcinoma cell lines. Oncotarget 2017, 8, 11676–11691. [Google Scholar] [CrossRef] [Green Version]
- Foreman, K.E.; Jesse, J.N.; Kuo, P.C.; Gupta, G.N. Emetine dihydrochloride: A novel therapy for bladder cancer. J. Urol. 2014, 191, 502–509. [Google Scholar] [CrossRef]
- Yosifov, D.Y.; Idler, I.; Bhattacharya, N.; Reichenzeller, M.; Close, V.; Ezerina, D.; Scheffold, A.; Jebaraj, B.M.C.; Kugler, S.; Bloehdorn, J.; et al. Oxidative stress as candidate therapeutic target to overcome microenvironmental protection of CLL. Leukemia 2019. [Google Scholar] [CrossRef]
- Mill, C.P.; Fiskus, W.; DiNardo, C.D.; Qian, Y.; Raina, K.; Rajapakshe, K.; Perera, D.; Coarfa, C.; Kadia, T.M.; Khoury, J.D.; et al. RUNX1-targeted therapy for AML expressing somatic or germline mutation in RUNX1. Blood 2019, 134, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Goyama, S.; Schibler, J.; Cunningham, L.; Zhang, Y.; Rao, Y.; Nishimoto, N.; Nakagawa, M.; Olsson, A.; Wunderlich, M.; Link, K.A.; et al. Transcription factor RUNX1 promotes survival of acute myeloid leukemia cells. J. Clin. Investig. 2013, 123, 3876–3888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrow, M.; Horton, S.; Kioussis, D.; Brady, H.J.; Williams, O. TEL-AML1 promotes development of specific hematopoietic lineages consistent with preleukemic activity. Blood 2004, 103, 3890–3896. [Google Scholar] [CrossRef] [PubMed]
- Vormoor, B.; Knizia, H.K.; Batey, M.A.; Almeida, G.S.; Wilson, I.; Dildey, P.; Sharma, A.; Blair, H.; Hide, I.G.; Heidenreich, O.; et al. Development of a preclinical orthotopic xenograft model of ewing sarcoma and other human malignant bone disease using advanced in vivo imaging. PLoS ONE 2014, 9, e85128. [Google Scholar] [CrossRef]
- Roulston, G.D.; Burt, C.L.; Kettyle, L.M.; Matchett, K.B.; Keenan, H.L.; Mulgrew, N.M.; Ramsey, J.M.; Dougan, C.; McKiernan, J.; Grishagin, I.V.; et al. Low-dose salinomycin induces anti-leukemic responses in AML and MLL. Oncotarget 2016, 7, 73448–73461. [Google Scholar] [CrossRef] [Green Version]
- Dunning, M.J.; Smith, M.L.; Ritchie, M.E.; Tavare, S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics 2007, 23, 2183–2184. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kettyle, L.M.; Lebert-Ghali, C.-É.; Grishagin, I.V.; Dickson, G.J.; O’Reilly, P.G.; Simpson, D.A.; Bijl, J.J.; Mills, K.I.; Sauvageau, G.; Thompson, A. Pathways, Processes, and Candidate Drugs Associated with a Hoxa Cluster-Dependency Model of Leukemia. Cancers 2019, 11, 2036. https://doi.org/10.3390/cancers11122036
Kettyle LM, Lebert-Ghali C-É, Grishagin IV, Dickson GJ, O’Reilly PG, Simpson DA, Bijl JJ, Mills KI, Sauvageau G, Thompson A. Pathways, Processes, and Candidate Drugs Associated with a Hoxa Cluster-Dependency Model of Leukemia. Cancers. 2019; 11(12):2036. https://doi.org/10.3390/cancers11122036
Chicago/Turabian StyleKettyle, Laura M., Charles-Étienne Lebert-Ghali, Ivan V. Grishagin, Glenda J. Dickson, Paul G. O’Reilly, David A. Simpson, Janet J. Bijl, Ken I. Mills, Guy Sauvageau, and Alexander Thompson. 2019. "Pathways, Processes, and Candidate Drugs Associated with a Hoxa Cluster-Dependency Model of Leukemia" Cancers 11, no. 12: 2036. https://doi.org/10.3390/cancers11122036