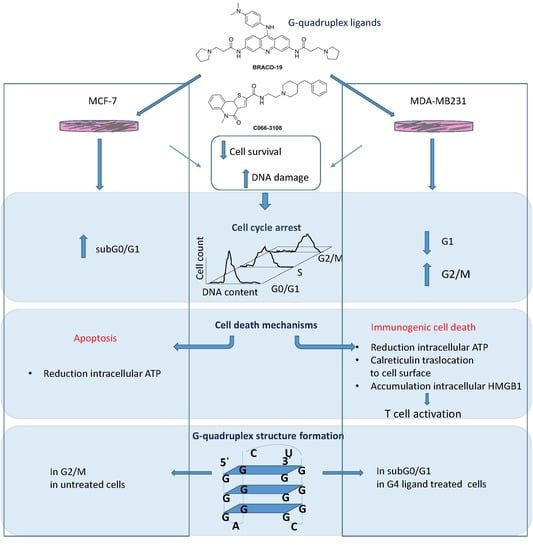
G-Quadruplex Binders Induce Immunogenic Cell Death Markers in Aggressive Breast Cancer Cells
Abstract
:1. Introduction
2. Results
2.1. Anti-Proliferative Effects of G4 Ligands in Cancer Cells
2.2. C066-3108 and BRACO-19 Induce DNA Damage in Breast Cancer Cells
2.3. C066-3108 and BRACO-19 Arrest Cell Cycle Progression
2.4. Apoptosis Induction in MCF-7 Cells by Flow Cytometry
2.5. Increase of Calreticulin Surface Exposure in MDA-MB-231 Cells
2.6. C066-3108 and BRACO-19 Decrease Intracellular ATP Release
2.7. C066-3108 and BRACO-19 Effects on Intracellular HMGB1
2.8. BRACO-19 Induces T-Cell Activation
2.9. G4 Structure Induction/Stabilization in Breast Cancer Cells
3. Discussion
4. Materials and Methods
4.1. Cells and Drugs
4.2. Sulforhodamine B (SRB) and MTT Proliferation Assays
4.3. γH2AX and/or G4 and PI Intracellular Staining
4.4. Cell cycle, Annexin V/PI and Calreticulin Staining
4.5. ATP Intracellular Concentration
4.6. HMGB1 Intracellular Staining
4.7. T-cell Activation Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| G-quadruplex | G4 |
| High mobility group box 1 | HMGB1 |
| Immunogenic cell death | ICD |
| Estrogen receptor | ER |
| Progesterone receptor | PR |
| Human epidermal growth factor receptor 2 | HER2 |
| Damage-associated molecular patterns | DAMPS |
| Peripheral blood mononuclear cells | PBMC |
| Sulforhodamine B | SRB |
| Phytohemagglutinin | PHA |
| Conditioned media | CM |
| Propidium iodide | PI |
| Telomere repeat binding factor 1 | TRF1 |
References
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef] [PubMed]
- Dexheimer, T.S.; Sun, D.; Hurley, L.H. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the Bcl-2 P1 promoter. J. Am. Chem. Soc. 2006, 128, 5404–5415. [Google Scholar] [CrossRef] [PubMed]
- Cogoi, S.; Xodo, L.E. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006, 34, 2536–2549. [Google Scholar] [CrossRef] [PubMed]
- Castor, K.J.; Liu, Z.; Fakhoury, J.; Hancock, M.A.; Mittermaier, A.; Moitessier, N.; Sleiman, H.F. A platinum(II) phenylphenanthroimidazole with an extended side-chain exhibits slow dissociation from a c-Kit G-quadruplex motif. Chemistry 2013, 19, 17836–17845. [Google Scholar] [CrossRef] [PubMed]
- Tauchi, T.; Shin-Ya, K.; Sashida, G.; Sumi, M.; Nakajima, A.; Shimamoto, T.; Ohyashiki, J.H.; Ohyashiki, K. Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: Involvement of ATM-dependent DNA damage response pathways. Oncogene 2003, 22, 5338–5347. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Xiang, J.F.; Yang, Q.F.; Sun, H.X.; Guan, A.J.; Tang, Y.L. G4ldb: A database for discovering and studying G-quadruplex ligands. Nucleic Acids Res. 2013, 41, D1115–D1123. [Google Scholar] [CrossRef]
- Monchaud, D.; Teulade-Fichou, M.P. A hitchhiker’s guide to G-quadruplex ligands. Org. Biomol. Chem. 2008, 6, 627–636. [Google Scholar] [CrossRef]
- Maji, B.; Bhattacharya, S. Advances in the molecular design of potential anticancer agents via targeting of human telomeric DNA. Chem. Commun. 2014, 50, 6422–6438. [Google Scholar] [CrossRef]
- Pennarun, G.; Granotier, C.; Gauthier, L.R.; Gomez, D.; Hoffschir, F.; Mandine, E.; Riou, J.F.; Mergny, J.L.; Mailliet, P.; Boussin, F.D. Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene 2005, 24, 2917–2928. [Google Scholar] [CrossRef]
- Rodriguez, R.; Miller, K.M.; Forment, J.V.; Bradshaw, C.R.; Nikan, M.; Britton, S.; Oelschlaegel, T.; Xhemalce, B.; Balasubramanian, S.; Jackson, S.P. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012, 8, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.X.; Su, H.F.; Lv, P.; Ma, Y.; Wang, S.K.; Miao, H.; Liu, H.Y.; Tan, J.H.; Ou, T.M.; Gu, L.Q.; et al. A newly identified berberine derivative induces cancer cell senescence by stabilizing endogenous G-quadruplexes and sparking a DNA damage response at the telomere region. Oncotarget 2015, 6, 35625–35635. [Google Scholar] [CrossRef] [PubMed]
- Gluszynska, A.; Juskowiak, B.; Rubis, B. Binding study of the fluorescent carbazole derivative with human telomeric G-quadruplexes. Molecules 2018, 23, 3154. [Google Scholar] [CrossRef] [PubMed]
- Kaulage, M.H.; Maji, B.; Pasadi, S.; Ali, A.; Bhattacharya, S.; Muniyappa, K. Targeting G-quadruplex DNA structures in the telomere and oncogene promoter regions by benzimidazolecarbazole ligands. Eur. J. Med. Chem. 2018, 148, 178–194. [Google Scholar] [CrossRef]
- Xu, H.; di Antonio, M.; McKinney, S.; Mathew, V.; Ho, B.; O’Neil, N.J.; Santos, N.D.; Silvester, J.; Wei, V.; Garcia, J.; et al. Cx-5461 is a DNA G-quadruplex stabilizer with selective lethality in Brca1/2 deficient tumours. Nat. Commun. 2017, 8, 14432. [Google Scholar] [CrossRef]
- Brooks, T.A.; Hurley, L.H. Targeting MYC expression through G-quadruplexes. Genes Cancer 2010, 1, 641–649. [Google Scholar] [CrossRef]
- Read, M.; Harrison, R.J.; Romagnoli, B.; Tanious, F.A.; Gowan, S.H.; Reszka, A.P.; Wilson, W.D.; Kelland, L.R.; Neidle, S. Structure-based design of selective and potent G quadruplex-mediated telomerase inhibitors. Proc. Natl. Acad. Sci. USA 2001, 98, 4844–4849. [Google Scholar] [CrossRef]
- Harrison, R.J.; Cuesta, J.; Chessari, G.; Read, M.A.; Basra, S.K.; Reszka, A.P.; Morrell, J.; Gowan, S.M.; Incles, C.M.; Tanious, F.A.; et al. Trisubstituted acridine derivatives as potent and selective telomerase inhibitors. J. Med. Chem. 2003, 46, 4463–4476. [Google Scholar] [CrossRef]
- Gowan, S.M.; Harrison, J.R.; Patterson, L.; Valenti, M.; Read, M.A.; Neidle, S.; Kelland, L.R. A G-quadruplex-interactive potent small-molecule inhibitor of telomerase exhibiting in vitro and in vivo antitumor activity. Mol. Pharmacol. 2002, 61, 1154–1162. [Google Scholar] [CrossRef]
- Zhou, G.; Liu, X.; Li, Y.; Xu, S.; Ma, C.; Wu, X.; Cheng, Y.; Yu, Z.; Zhao, G.; Chen, Y. Telomere targeting with a novel G-quadruplex-interactive ligand braco-19 induces T-loop disassembly and telomerase displacement in human glioblastoma cells. Oncotarget 2016, 7, 14925–14939. [Google Scholar] [CrossRef]
- di Leva, F.S.; Zizza, P.; Cingolani, C.; D’Angelo, C.; Pagano, B.; Amato, J.; Salvati, E.; Sissi, C.; Pinato, O.; Marinelli, L.; et al. Exploring the chemical space of G-quadruplex binders: Discovery of a novel chemotype targeting the human telomeric sequence. J. Med. Chem. 2013, 56, 9646–9654. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Buque, A.; Kepp, O.; Zitvogel, L.; Kroemer, G. Immunogenic cell death in cancer and infectious disease. Nat. Rev. Immunol. 2017, 17, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Mouw, K.W.; Goldberg, M.S.; Konstantinopoulos, P.A.; D’Andrea, A.D. DNA damage and repair biomarkers of immunotherapy response. Cancer Discov. 2017, 7, 675–693. [Google Scholar] [CrossRef] [PubMed]
- Rufo, N.; Garg, A.D.; Agostinis, P. The unfolded protein response in immunogenic cell death and cancer immunotherapy. Trends Cancer 2017, 3, 643–658. [Google Scholar] [CrossRef] [PubMed]
- Yatim, N.; Cullen, S.; Albert, M.L. Dying cells actively regulate adaptive immune responses. Nat. Rev. Immunol. 2017, 17, 262–275. [Google Scholar] [CrossRef] [PubMed]
- Nakhjavani, M.; Hardingham, J.E.; Palethorpe, H.M.; Price, T.J.; Townsend, A.R. Druggable molecular targets for the treatment of triple negative breast cancer. J. Breast Cancer 2019, 22, 341–361. [Google Scholar] [CrossRef] [PubMed]
- Laezza, C.; Malfitano, A.M.; di Matola, T.; Ricchi, P.; Bifulco, M. Involvement of Akt/Nf-κB pathway in N6-isopentenyladenosine-induced apoptosis in human breast cancer cells. Mol. Carcinog. 2010, 49, 892–901. [Google Scholar] [CrossRef]
- Amato, J.; Madanayake, T.W.; Iaccarino, N.; Novellino, E.; Randazzo, A.; Hurley, L.H.; Pagano, B. Hmgb1 binds to the KRAS promoter G-quadruplex: A new player in oncogene transcriptional regulation? Chem. Commun. 2018, 54, 9442–9445. [Google Scholar] [CrossRef]
- Amato, J.; Cerofolini, L.; Brancaccio, D.; Giuntini, S.; Iaccarino, N.; Zizza, P.; Iachettini, S.; Biroccio, A.; Novellino, E.; Rosato, A.; et al. Insights into Telomeric G-Quadruplex DNA Recognition by Hmgb1 Protein. Nucleic Acids Res. 2019, 47, 9950–9966. [Google Scholar] [CrossRef]
- Debray, J.; Zeghida, W.; Jourdan, M.; Monchaud, D.; Dheu-Andries, M.L.; Dumy, P.; Teulade-Fichou, M.P.; Demeunynck, M. Synthesis and evaluation of fused bispyrimidinoacridines as novel pentacyclic analogues of quadruplex-binder braco-19. Org. Biomol. Chem. 2009, 7, 5219–5228. [Google Scholar] [CrossRef]
- Golden, E.B.; Frances, D.; Pellicciotta, I.; Demaria, S.; Barcellos-Hoff, M.H.; Formenti, S.C. Radiation fosters dose-dependent and chemotherapy-induced immunogenic cell death. Oncoimmunology 2014, 3, e28518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, R.; Chen, R.; Zhang, Q.; Hou, W.; Wu, S.; Cao, L.; Huang, J.; Yu, Y.; Fan, X.G.; Yan, Z.; et al. Hmgb1 in health and disease. Mol. Asp. Med. 2014, 40, 1–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pagano, B.; Amato, J.; Iaccarino, N.; Cingolani, C.; Zizza, P.; Biroccio, A.; Novellino, E.; Randazzo, A. Looking for efficient G-quadruplex ligands: Evidence for selective stabilizing properties and telomere damage by drug-like molecules. ChemMedChem 2015, 10, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Hato, S.V.; Khong, A.; de Vries, I.J.; Lesterhuis, W.J. Molecular pathways: The immunogenic effects of platinum-based chemotherapeutics. Clin. Cancer Res. 2014, 20, 2831–2837. [Google Scholar] [CrossRef]
- Ren, J.; Chaires, J.B. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry 1999, 38, 16067–16075. [Google Scholar] [CrossRef]
- Wang, A.H.; Ughetto, G.; Quigley, G.J.; Rich, A. Interactions between an anthracycline antibiotic and DNA: Molecular structure of daunomycin complexed to d(CpGpTpApCpG) at 1.2-a resolution. Biochemistry 1987, 26, 1152–1163. [Google Scholar] [CrossRef]
- Malfitano, A.M.; Sosa, S.; Laezza, C.; de Bortoli, M.; Tubaro, A.; Bifulco, M. Rimonabant reduces keratinocyte viability by induction of apoptosis and exerts topical anti-inflammatory activity in mice. Br. J. Pharmacol. 2011, 162, 84–93. [Google Scholar] [CrossRef] [Green Version]
- Di Natale, C.; la Manna, S.; Malfitano, A.M.; di Somma, S.; Florio, D.; Scognamiglio, P.L.; Novellino, E.; Netti, P.A.; Marasco, D. Structural insights into amyloid structures of the C-terminal region of nucleophosmin 1 in type A mutation of acute myeloid leukemia. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 637–644. [Google Scholar] [CrossRef]










| G4 Ligand Effects | MCF-7 | MDA-MB231 |
|---|---|---|
| Anti-proliferative effects | √ | √ |
| yH2AX phosphorylation | √ | √ |
| Apoptosis | √ | x |
| Immunogenic cell death markers: Reduction of ATP Calreticulin exposure Enhanced intracellular HMGB1 | √ x x | √ √ √ |
| T-cell activation | not investigated | √ |
| G4 structure induction | x | √ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Somma, S.; Amato, J.; Iaccarino, N.; Pagano, B.; Randazzo, A.; Portella, G.; Malfitano, A.M. G-Quadruplex Binders Induce Immunogenic Cell Death Markers in Aggressive Breast Cancer Cells. Cancers 2019, 11, 1797. https://doi.org/10.3390/cancers11111797
Di Somma S, Amato J, Iaccarino N, Pagano B, Randazzo A, Portella G, Malfitano AM. G-Quadruplex Binders Induce Immunogenic Cell Death Markers in Aggressive Breast Cancer Cells. Cancers. 2019; 11(11):1797. https://doi.org/10.3390/cancers11111797
Chicago/Turabian StyleDi Somma, Sarah, Jussara Amato, Nunzia Iaccarino, Bruno Pagano, Antonio Randazzo, Giuseppe Portella, and Anna Maria Malfitano. 2019. "G-Quadruplex Binders Induce Immunogenic Cell Death Markers in Aggressive Breast Cancer Cells" Cancers 11, no. 11: 1797. https://doi.org/10.3390/cancers11111797