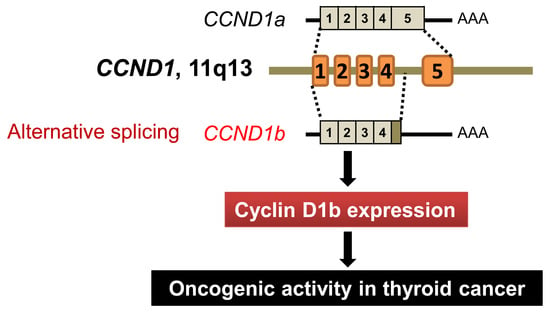
CCND1 Splice Variant as A Novel Diagnostic and Predictive Biomarker for Thyroid Cancer
Abstract
:1. Introduction
2. Results
2.1. CCND1 G870A Polymorphism (rs9344) and CCND1 mRNA Expression
2.2. Relationship between the mRNA Expression of CCND1 Isoforms and Types of Thyroid Tumor
2.3. Protein Expression of Cyclin D1 Isoforms in Different Types of Thyroid Tumors
2.4. Clinical Impact of Expression of CCND1 mRNA Isoforms and Cyclin D1b Protein in PTC
2.5. Expression of CCND1b mRNA and Cyclin D1b Protein in NIFTP and Invasive Encapsulated Follicular Variant of PTC
2.6. A Case of Noninvasive Encapsulated PTC with Predominant Follicular Growth and BRAF V600E Mutation
2.7. CCND1 Mutation and mRNA Expression in TCGA Dataset
3. Discussion
4. Materials and Methods
4.1. Patient and Clinical Samples
4.2. Isolation of Nucleic Acids
4.3. Genotyping of CCND1 G870A Polymorphism
4.4. Quantitative Real-Time PCR for CCND1 Alternative Transcripts
4.5. Molecular Analysis of BRAF, NRAS, HRAS, and KRAS Genes
4.6. Antibody Preparation
4.7. Western Blotting for Assessing Antibody Specificity
4.8. Immunohistochemistry
4.9. The Cancer Genome Atlas Data Analysis for CCND1
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nikiforov, Y.E.; Baloch, Z.W.; Belge, G.; Chan, J.K.C.; Derwahl, K.M.; Evans, H.L.; Fagin, J.A.; Ghossein, R.A.; Lloyd, R.V.; Oriola, J.; et al. Tumors of the thyroid gland. In WHO Classification of Tumours of Endocrine Organs; Lloyd, R.V., Osamura, R.Y., Klöppel, G., Rosai, J., Eds.; WHO Press: Geneva, Switzerland, 2017; Volume 10, pp. 66–103. [Google Scholar]
- Nikiforov, Y.E.; Seethala, R.R.; Tallini, G.; Baloch, Z.W.; Basolo, F.; Thompson, L.D.; Barletta, J.A.; Wenig, B.M.; Al Ghuzlan, A.; Kakudo, K.; et al. Nomenclature Revision for Encapsulated Follicular Variant of Papillary Thyroid Carcinoma: A Paradigm Shift to Reduce Overtreatment of Indolent Tumors. JAMA Oncol. 2016, 2, 1023–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, B.Y.; Choi, H.S.; Park, Y.J.; Lim, J.A.; Ahn, H.Y.; Lee, E.K.; Kim, K.W.; Yi, K.H.; Chung, J.K.; Youn, Y.K.; et al. Changes in the clinicopathological characteristics and outcomes of thyroid cancer in Korea over the past four decades. Thyroid 2013, 23, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.K.; Little, M.P.; Lubin, J.H.; Brenner, A.V.; Wells, S.A., Jr.; Sigurdson, A.J.; Nikiforov, Y.E. The increase in thyroid cancer incidence during the last four decades is accompanied by a high frequency of BRAF mutations and a sharp increase in RAS mutations. J. Clin. Endocrinol. Metab. 2014, 99, E276–E285. [Google Scholar] [CrossRef] [PubMed]
- Ahn, H.S.; Kim, H.J.; Welch, H.G. Korea’s thyroid-cancer “epidemic”—Screening and overdiagnosis. N. Engl. J. Med. 2014, 371, 1765–1767. [Google Scholar] [CrossRef] [PubMed]
- Vaccarella, S.; Franceschi, S.; Bray, F.; Wild, C.P.; Plummer, M.; Dal Maso, L. Worldwide thyroid-cancer epidemic? The increasing impact of overdiagnosis. N. Engl. J. Med. 2016, 375, 614–617. [Google Scholar] [CrossRef] [PubMed]
- Ahn, H.S.; Kim, H.J.; Kim, K.H.; Lee, Y.S.; Han, S.J.; Kim, Y.; Ko, M.J.; Brito, J.P. Thyroid cancer screening in South Korea increases detection of papillary cancers with no impact on other subtypes or thyroid cancer mortality. Thyroid 2016, 26, 1535–1540. [Google Scholar] [CrossRef] [PubMed]
- Fagin, J.A.; Wells, S.A., Jr. Biologic and clinical perspectives on thyroid cancer. N. Engl. J. Med. 2016, 375, 1054–1067. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, K.E.; Diehl, J.A.; Haiman, C.A.; Knudsen, E.S. Cyclin D1: Polymorphism, aberrant splicing and cancer risk. Oncogene 2006, 25, 1620–1628. [Google Scholar] [CrossRef] [PubMed]
- Alao, J.P. The regulation of cyclin D1 degradation: Roles in cancer development and the potential for therapeutic invention. Mol. Cancer 2007, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.K.; Kang, Y.G.; Bae, J.S.; Lim, D.J.; Choi, Y.J.; Lee, K.Y. Unique patterns of tumor growth related with the risk of lymph node metastasis in papillary thyroid carcinoma. Mod. Pathol. 2010, 23, 1201–1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, N.; Akbani, R.; Aksoy, B.A.; Ally, A.; Arachchi, H.; Asa, S.L.; Auman, J.T.; Balasundaram, M.; Balu, S.; Baylin, S.B.; et al. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014, 159, 676–690. [Google Scholar] [CrossRef] [PubMed]
- Landa, I.; Ibrahimpasic, T.; Boucai, L.; Sinha, R.; Knauf, J.A.; Shah, R.H.; Dogan, S.; Ricarte-Filho, J.C.; Krishnamoorthy, G.P.; Xu, B.; et al. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J. Clin. Investig. 2016, 126, 1052–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comstock, C.E.; Augello, M.A.; Benito, R.P.; Karch, J.; Tran, T.H.; Utama, F.E.; Tindall, E.A.; Wang, Y.; Burd, C.J.; Groh, E.M.; et al. Cyclin D1 splice variants: Polymorphism, risk, and isoform-specific regulation in prostate cancer. Clin. Cancer Res. 2009, 15, 5338–5349. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.J.; Tambe, Y.; Mukaisho, K.; Sugihara, H.; Isono, T.; Sonoda, H.; Shimizu, T.; Kondoh, G.; Inoue, H. Female-specific rectal carcinogenesis in cyclin D1b transgenic mice. Carcinogenesis 2014, 35, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, G.; Bittencourt, D.; Laud, K.; Barbier, J.; Delattre, O.; Auboeuf, D.; Dutertre, M. Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer. Proc. Natl. Acad. Sci. USA 2008, 105, 6004–6009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.J.; Nishi, K.; Isono, T.; Okuyama, Y.; Tambe, Y.; Okada, Y.; Inoue, H. Cyclin D1b variant promotes cell invasiveness independent of binding to CDK4 in human bladder cancer cells. Mol. Carcinog. 2009, 48, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Millar, E.K.; Dean, J.L.; McNeil, C.M.; O’Toole, S.A.; Henshall, S.M.; Tran, T.; Lin, J.; Quong, A.; Comstock, C.E.; Witkiewicz, A.; et al. Cyclin D1b protein expression in breast cancer is independent of cyclin D1a and associated with poor disease outcome. Oncogene 2009, 28, 1812–1820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; An, S.J.; Chen, Z.H.; Zhang, G.C.; Zhu, J.Q.; Nie, Q.; Xie, Z.; Guo, A.L.; Mok, T.S.; Wu, Y.L. Expression of cyclin D1 splice variants is differentially associated with outcome in non-small cell lung cancer patients. Hum. Pathol. 2008, 39, 1792–1801. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; Feber, A.; Xi, L.; Pennathur, A.; Wu, M.; Luketich, J.D.; Godfrey, T.E. Association between CCND1 G/A870 polymorphism, allele-specific amplification, cyclin D1 expression, and survival in esophageal and lung carcinoma. Clin. Cancer Res. 2008, 14, 7804–7812. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Eu, K.W.; Seow-Choen, F.; Fook-Chong, S.; Cheah, P.Y. GG genotype of cyclin D1 G870A polymorphism is associated with increased risk and advanced colorectal cancer in patients in Singapore. Eur. J. Cancer 2005, 41, 1037–1044. [Google Scholar] [CrossRef] [PubMed]
- Shakir, R.; Ngo, N.; Naresh, K.N. Correlation of cyclin D1 transcript levels, transcript type and protein expression with proliferation and histology among mantle cell lymphoma. J. Clin. Pathol. 2008, 61, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Zeybek, U.; Yaylim, I.; Ozkan, N.E.; Korkmaz, G.; Turan, S.; Kafadar, D.; Cacina, C.; Kafadar, A.M. Cyclin D1 gene G870A variants and primary brain tumors. Asian Pac. J. Cancer Prev. 2013, 14, 4101–4106. [Google Scholar] [CrossRef] [PubMed]
- Jeyapalan, J.N.; Doctor, G.T.; Jones, T.A.; Alberman, S.N.; Tep, A.; Haria, C.M.; Schwalbe, E.C.; Morley, I.C.; Hill, A.A.; LeCain, M.; et al. DNA methylation analysis of paediatric low-grade astrocytomas identifies a tumour-specific hypomethylation signature in pilocytic astrocytomas. Acta. Neuropathol. Commun. 2016, 4, 54. [Google Scholar] [CrossRef] [PubMed]
- Abramson, V.G.; Troxel, A.B.; Feldman, M.; Mies, C.; Wang, Y.; Sherman, L.; McNally, S.; Diehl, A.; Demichele, A. Cyclin D1b in human breast carcinoma and coexpression with cyclin D1a is associated with poor outcome. Anticancer Res. 2010, 30, 1279–1285. [Google Scholar] [PubMed]
- Nikiforov, Y.E.; Baloch, Z.W.; Hodak, S.P.; Giordano, T.J.; Lloyd, R.V.; Seethala, R.R.; Wenig, B.M. Change in Diagnostic Criteria for Noninvasive Follicular Thyroid Neoplasm With Papillarylike Nuclear Features. JAMA Oncol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Hryhorowicz, S.; Ziemnicka, K.; Kaczmarek-RyŚ, M.; Hoppe-GoŁĘBiewska, J.; PŁAwski, A.; Skrzypczak-ZieliŃSka, M.; Szkudlarek, M.; GoŁĄB, M.; Budny, B.; RuchaŁA, M.; et al. CCND1 gene polymorphic variants in patients with differentiated thyroid carcinoma. Oncol Lett. 2015, 9, 442–448. [Google Scholar] [CrossRef] [PubMed]
- Aytekin, T.; Aytekin, A.; Maralcan, G.; Gokalp, M.A.; Ozen, D.; Borazan, E.; Yilmaz, L. A cyclin D1 (CCND1) gene polymorphism contributes to susceptibility to papillary thyroid cancer in the Turkish population. Asian Pac. J. Cancer Prev. 2014, 15, 7181–7185. [Google Scholar] [CrossRef] [PubMed]
- Pešutić-Pisac, V.; Punda, A.; Glunčić, I.; Bedeković, V.; Pranić-Kragić, A.; Kunac, N. Cyclin D1 and p27 expression as prognostic factor in papillary carcinoma of the thyroid: Association with clinicopathological parameters. Croat Med. J. 2008, 49, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Khoo, M.L.; Ezzat, S.; Freeman, J.L.; Asa, S.L. Cyclin D1 protein expression predicts metastatic behavior in thyroid papillary microcarcinomas but is not associated with gene amplification. J. Clin. Endocrinol. Metab. 2002, 87, 1810–1813. [Google Scholar] [CrossRef] [PubMed]
- Seybt, T.P.; Ramalingam, P.; Huang, J.; Looney, S.W.; Reid, M.D. Cyclin D1 expression in benign and differentiated malignant tumors of the thyroid gland: Diagnostic and biologic implications. Appl. Immunohistochem. Mol. Morphol. 2012, 20, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.J.; Terado, T.; Tambe, Y.; Mukaisho, K.I.; Sugihara, H.; Kawauchi, A.; Inoue, H. Anti-oncogenic activities of cyclin D1b siRNA on human bladder cancer cells via induction of apoptosis and suppression of cancer cell stemness and invasiveness. Int. J. Oncol. 2018, 52, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Fuste, N.P.; Fernandez-Hernandez, R.; Cemeli, T.; Mirantes, C.; Pedraza, N.; Rafel, M.; Torres-Rosell, J.; Colomina, N.; Ferrezuelo, F.; Dolcet, X.; et al. Cytoplasmic cyclin D1 regulates cell invasion and metastasis through the phosphorylation of paxillin. Nat. Commun. 2016, 7, 11581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, D.I.; Barbash, O.; Kumar, K.G.; Weber, J.D.; Harper, J.W.; Klein-Szanto, A.J.; Rustgi, A.; Fuchs, S.Y.; Diehl, J.A. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol. Cell. 2006, 24, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Fuste, N.P.; Castelblanco, E.; Felip, I.; Santacana, M.; Fernandez-Hernandez, R.; Gatius, S.; Pedraza, N.; Pallares, J.; Cemeli, T.; Valls, J.; et al. Characterization of cytoplasmic cyclin D1 as a marker of invasiveness in cancer. Oncotarget 2016, 7, 26979–26991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Body, S.; Esteve-Arenys, A.; Miloudi, H.; Recasens-Zorzo, C.; Tchakarska, G.; Moros, A.; Bustany, S.; Vidal-Crespo, A.; Rodriguez, V.; Lavigne, R.; et al. Cytoplasmic cyclin D1 controls the migration and invasiveness of mantle lymphoma cells. Sci. Rep. 2017, 7, 13946. [Google Scholar] [CrossRef] [PubMed]
- Cho, U.; Mete, O.; Kim, M.H.; Bae, J.S.; Jung, C.K. Molecular correlates and rate of lymph node metastasis of non-invasive follicular thyroid neoplasm with papillary-like nuclear features and invasive follicular variant papillary thyroid carcinoma: The impact of rigid criteria to distinguish non-invasive follicular thyroid neoplasm with papillary-like nuclear features. Mod. Pathol. 2017, 30, 810–825. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.E.; Hwang, T.S.; Choi, Y.L.; Kim, W.Y.; Han, H.S.; Lim, S.D.; Kim, W.S.; Yoo, Y.B.; Kim, S.K. Molecular profiling of papillary thyroid carcinoma in Korea with a high prevalence of BRAF(V600E) Mutation. Thyroid 2017, 27, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Parente, D.N.; Kluijfhout, W.P.; Bongers, P.J.; Verzijl, R.; Devon, K.M.; Rotstein, L.E.; Goldstein, D.P.; Asa, S.L.; Mete, O.; Pasternak, J.D. Clinical safety of renaming encapsulated follicular variant of papillary thyroid carcinoma: Is NIFTP truly benign? World J. Surg. 2018, 42, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.S.; Harrison, G.P.; Datto, M.B. Young Investigator Challenge: Molecular testing in noninvasive follicular thyroid neoplasm with papillary-like nuclear features. Cancer Cytopathol. 2016, 124, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Wang, X.; Huang, X.; Gui, H.; Li, Y.; Chen, Q.; Liu, D.; Liu, L. Diagnostic significance of CK19, galectin-3, CD56, TPO and Ki67 expression and BRAF mutation in papillary thyroid carcinoma. Oncol Lett. 2018, 15, 4269–4277. [Google Scholar] [CrossRef] [PubMed]
- Nasr, M.R.; Mukhopadhyay, S.; Zhang, S.; Katzenstein, A.L. Absence of the BRAF mutation in HBME1+ and CK19+ atypical cell clusters in Hashimoto thyroiditis: Supportive evidence against preneoplastic change. Am. J. Clin. Pathol. 2009, 132, 906–912. [Google Scholar] [CrossRef] [PubMed]
- Kashofer, K.; Viertler, C.; Pichler, M.; Zatloukal, K. Quality control of RNA preservation and extraction from paraffin-embedded tissue: Implications for RT-PCR and microarray analysis. PLoS ONE 2013, 8, e70714. [Google Scholar] [CrossRef] [PubMed]
- Haugen, B.R.; Alexander, E.K.; Bible, K.C.; Doherty, G.M.; Mandel, S.J.; Nikiforov, Y.E.; Pacini, F.; Randolph, G.W.; Sawka, A.M.; Schlumberger, M.; et al. 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer: The American Thyroid Association Guidelines Task Force on Thyroid Nodules and Differentiated Thyroid Cancer. Thyroid 2016, 26, 1–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Tumor Type | n | Cyclin D1a | Cyclin D1b | |
|---|---|---|---|---|
| Nuclear | Nuclear | Cytoplasmic | ||
| Cohort 1 | 281 | |||
| Nodular hyperplasia | 16 | 0 | 0 | 0 |
| Follicular adenoma | 33 | 8 (33%) | 0 | 0 |
| NIFTP | 9 | 9 (100%) | 1 (11%) | 0 |
| Papillary thyroid carcinoma | 170 | 170 (100%) | 110 (64.7%) | 124 (72.9%) |
| Classic | 125 | 125 (100%) | 83 (66.4%) | 93 (74.4%) |
| Follicular variant | 12 | 12 (100%) | 3 (25%) | 3 (25%) |
| Tall cell variant | 33 | 33 (100%) | 24 (73%) | 28 (85%) |
| Follicular thyroid carcinoma | 32 | 32 (100%) | 14 (13%) | 15 (16%) |
| Poorly differentiated thyroid carcinoma | 4 | 4 (100%) | 2 (50%) | 4 (100%) |
| Medullary thyroid carcinoma | 17 | 17 (100%) | 12 (71%) | 12 (71%) |
| Cohort 2 | 58 | |||
| NIFTP | 34 | 34 (100%) | 5 (15%) | 4 (12%) |
| Invasive encapsulated follicular variant of papillary thyroid carcinoma | 24 | 24 (100%) | 9 (38%) | 7 (29%) |
| Characteristic | n | High Expression of CCND1 mRNA Isoforms | High Expression of Cyclin D1b Protein | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CCND1a | p-Value | CCND1b | p-Value | Nuclear | p-Value | Cytoplasmic | p-Value | ||
| Age (years) | 0.042 | 0.660 | 0.422 | 0.801 | |||||
| <55 | 127 | 57 (44.9%) | 63 (49.6%) | 80 (63.0%) | 92 (72.4%) | ||||
| ≥55 | 43 | 27 (62.8%) | 23 (53.5%) | 30 (69.8%) | 32 (74.4%) | ||||
| Gender | 0.396 | 0.916 | 0.423 | 0.208 | |||||
| Female | 133 | 68 (51.1%) | 67 (50.4%) | 84 (63.2%) | 94 (70.7%) | ||||
| Male | 37 | 16 (43.2%) | 19 (51.4%) | 26 (70.3%) | 30 (81.1%) | ||||
| Primary tumor size (cm) | 0.070 | 0.268 | 0.357 | 0.149 | |||||
| ≤1.0 | 93 | 40 (42.1%) | 43 (45.3%) | 60 (63.2%) | 66 (69.5%) | ||||
| 1.0–2.0 | 49 | 27 (55.1%) | 29 (59.2%) | 30 (61.2%) | 35 (71.4%) | ||||
| >2.0 | 26 | 17 (65.4%) | 14 (53.8%) | 20 (76.9%) | 23 (88.5%) | ||||
| Histologic variant | 0.000 | 0.668 | 0.009 | 0.000 | |||||
| Classic | 125 | 47 (37.6%) | 61 (48.8%) | 83 (66.4%) | 93 (74.4%) | ||||
| Follicular variant | 12 | 9 (75.0%) | 6 (50.0%) | 3 (25.0%) | 3 (25.0%) | ||||
| Tall cell variant | 33 | 28 (84.8%) | 19 (57.6%) | 24 (72.7%) | 28 (84.8%) | ||||
| Extrathyroidal extension | 0.219 | 0.633 | 0.516 | 0.150 | |||||
| Absent | 75 | 32 (42.7%) | 35 (46.7%) | 47 (62.7%) | 51 (68.0%) | ||||
| Microscopic | 81 | 43 (53.1%) | 44 (54.3%) | 52 (64.2%) | 60 (74.1%) | ||||
| Gross | 14 | 9 (64.3%) | 7 (50.0%) | 11 (78.6%) | 13 (92.9%) | ||||
| Lymph node metastasis | 0.638 | 0.047 | 0.002 | 0.000 | |||||
| Absent | 80 | 38 (47.5%) | 34 (42.5%) | 42 (52.5%) | 48 (60.0%) | ||||
| Present | 90 | 46 (51.1%) | 52 (57.8%) | 68 (75.6%) | 76 (84.4%) | ||||
| Lateral lymph node metastasis | 0.152 | 0.613 | 0.059 | 0.010 | |||||
| Absent | 137 | 64 (46.7%) | 68 (49.6%) | 84 (61.3%) | 94 (68.6%) | ||||
| Present | 33 | 20 (60.6%) | 18 (54.5%) | 26 (78.8%) | 30 (90.9%) | ||||
| Distant metastasis | 0.028 | 0.059 | 0.163 | 0.325 | |||||
| Absent | 165 | 79 (47.9%) | 81 (49.1%) | 105 (63.6%) | 119 (72.1%) | ||||
| Present | 5 | 5 (100%) | 5 (100%) | 5 (100%) | 5 (100%) | ||||
| BRAF V600E mutation | 0.276 | 0.893 | 0.108 | 0.323 | |||||
| Negative | 29 | 17 (58.6%) | 15 (51.7%) | 15 (51.7%) | 19 (65.5%) | ||||
| Positive | 141 | 67 (47.5%) | 71 (50.4%) | 95 (67.4%) | 105 (74.5%) | ||||
| Recurrence risk | 0.060 | 0.286 | 0.043 | 0.010 | |||||
| Low | 55 | 20 (36.4%) | 23 (41.8%) | 32 (58.2%) | 35 (63.6%) | ||||
| Intermediate | 84 | 46 (54.8%) | 46 (54.8%) | 52 (61.9%) | 60 (71.4%) | ||||
| High | 31 | 18 (58.1%) | 17 (54.8%) | 26 (83.9%) | 29 (93.5%) | ||||
| AJCC stage, 7th edition | 0.233 | 0.001 | 0.047 | 0.303 | |||||
| I | 97 | 44 (45.4%) | 39 (40.2%) | 57 (58.8%) | 66 (68.0%) | ||||
| II | 3 | 2 (66.7%) | 2 (66.7%) | 2 (66.7%) | 2 (66.7%) | ||||
| III | 67 | 35 (52.2%) | 42 (62.7%) | 48 (71.6%) | 53 (79.1%) | ||||
| IV | 3 | 3 (100%) | 3 (100%) | 3 (100%) | 3 (100%) | ||||
| AJCC stage, 8th edition | 0.022 | 0.011 | 0.081 | 0.282 | |||||
| I | 149 | 69 (46.3%) | 70 (47.0%) | 93 (62.4%) | 107 (71.8%) | ||||
| II | 19 | 13 (68.4%) | 14 (73.7%) | 15 (78.9%) | 15 (78.9%) | ||||
| IV | 2 | 2 (100%) | 2 (100%) | 2 (100%) | 2 (100%) | ||||
| Characteristic | n | High Expression of CCND1 mRNA Isoforms | High Expression of Cyclin D1b Protein | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CCND1a | p-Value | CCND1b | p-Value | Nuclear | p-Value | Cytoplasmic | p-Value | ||
| Age (years) | 0.022 | 0.438 | 0.289 | 0.358 | |||||
| <55 | 94 | 30 (31.9%) | 44 (46.8%) | 60 (63.8%) | 68 (72.3%) | ||||
| ≥55 | 31 | 17 (54.8%) | 17 (54.8%) | 23 (74.2%) | 25 (80.6%) | ||||
| Gender | 0.499 | 0.885 | 0.122 | 0.040 | |||||
| Female | 97 | 38 (39.2%) | 47 (48.5%) | 61 (62.9%) | 68 (70.1%) | ||||
| Male | 28 | 9 (32.1%) | 14 (50.0%) | 22 (78.6%) | 25 (89.3%) | ||||
| Primary tumor size (cm) | 0.709 | 0.078 | 0.473 | 0.156 | |||||
| ≤1.0 | 80 | 28 (35.0%) | 33 (41.3%) | 51 (63.8%) | 56 (70.0%) | ||||
| 1.0–2.0 | 30 | 13 (43.3%) | 19 (63.3%) | 20 (66.7%) | 23 (76.7%) | ||||
| >2.0 | 15 | 6 (40.0%) | 9 (60.0%) | 12 (80.0%) | 14 (93.3%) | ||||
| Extrathyroidal extension | 0.206 | 0.084 | 0.823 | 0.411 | |||||
| Absent | 59 | 19 (32.2%) | 24 (40.7%) | 38 (64.4%) | 41 (69.5%) | ||||
| Microscopic | 58 | 24 (41.4%) | 32 (55.2%) | 39 (67.2%) | 45 (77.6%) | ||||
| Gross | 8 | 4 (50.0%) | 5 (62.5%) | 6 (75.0%) | 7 (87.5%) | ||||
| Lymph node metastasis | 0.661 | 0.038 | 0.006 | 0.005 | |||||
| Absent | 59 | 21 (35.6%) | 23 (39.0%) | 32 (54.2%) | 37 (62.7%) | ||||
| Present | 66 | 26 (39.4%) | 38 (57.6%) | 51 (77.3%) | 56 (84.8%) | ||||
| Lateral lymph node metastasis | 0.142 | 0.144 | 0.202 | 0.065 | |||||
| Absent | 99 | 34 (34.3%) | 45 (45.5%) | 63 (63.6%) | 70 (70.7%) | ||||
| Present | 26 | 13 (50.0%) | 16 (61.5%) | 20 (76.9%) | 23 (88.5%) | ||||
| Distant metastasis | 0.007 | 0.059 | 0.167 | 0.327 | |||||
| Absent | 120 | 42 (35.0%) | 81 (49.1%) | 78 (65.0%) | 88 (73.3%) | ||||
| Present | 5 | 5 (100%) | 5 (100%) | 5 (100%) | 5 (100%) | ||||
| BRAF V600E mutation | 0.743 | 0.713 | 0.694 | 0.234 | |||||
| Negative | 17 | 7 (41.2%) | 9 (52.9%) | 12 (70.6%) | 15 (88.2%) | ||||
| Positive | 108 | 40 (37.0%) | 52 (48.1%) | 71 (65.7%) | 78 (72.2%) | ||||
| Recurrence risk | 0.258 | 0.056 | 0.190 | 0.111 | |||||
| Low | 48 | 14 (29.2%) | 17 (35.4%) | 30 (62.5%) | 33 (68.8%) | ||||
| Intermediate | 54 | 22 (40.7%) | 30 (55.6%) | 34 (63.0%) | 39 (72.2%) | ||||
| High | 23 | 11 (47.8%) | 14 (60.9%) | 19 (82.6%) | 21 (91.3%) | ||||
| AJCC stage, 7th edition | 0.014 | 0.007 | 0.065 | 0.025 | |||||
| I | 70 | 20 (28.6%) | 27 (38.6%) | 42 (60.0%) | 47 (67.1%) | ||||
| II | 3 | 2 (66.7%) | 2 (66.7%) | 2 (66.7%) | 2 (66.7%) | ||||
| III | 47 | 22 (44.9%) | 29 (59.2%) | 36 (73.5%) | 41 (83.7%) | ||||
| IV | 3 | 3 (100%) | 3 (100%) | 3 (100%) | 3 (100%) | ||||
| AJCC stage, 8th edition | 0.004 | 0.008 | 0.038 | 0.131 | |||||
| I | 107 | 35 (32.7%) | 47 (43.9%) | 67 (62.6%) | 77 (72.0%) | ||||
| II | 16 | 10 (62.5%) | 12 (75.0%) | 14 (87.5%) | 14 (87.5%) | ||||
| IV | 2 | 2 (100%) | 2 (100%) | 2 (100%) | 2 (100%) | ||||
| Molecular Alteration | NIFTP (n = 34) | Invasive Encapsulated Follicular Variant of Papillary Thyroid Carcinoma (n = 24) | p-Value |
|---|---|---|---|
| High expression of CCND1b mRNA | 18 (52.9%) | 13 (54.2%) | 0.927 |
| High expression of nuclear cyclin D1b | 5 (14.7%) | 9 (37.5%) | 0.046 |
| High expression of cytoplasmic cyclin D1b | 4 (11.8%) | 7 (29.2%) | 0.096 |
| BRAF V600E mutation | 0 | 0 | |
| BRAF K601E mutation | 2 (5.9%) | 0 | 0.339 |
| RAS mutation (total) | 20 (58.8%) | 13 (54.2%) | 0.724 |
| NRAS mutation | 14 (41.2%) | 8 (33.3%) | 0.544 |
| HRAS mutation | 6 (17.6%) | 4 (16.7%) | 0.605 |
| KRAS mutation | 0 | 2 (8.3%) | 0.167 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeon, S.; Kim, Y.; Jeong, Y.M.; Bae, J.S.; Jung, C.K. CCND1 Splice Variant as A Novel Diagnostic and Predictive Biomarker for Thyroid Cancer. Cancers 2018, 10, 437. https://doi.org/10.3390/cancers10110437
Jeon S, Kim Y, Jeong YM, Bae JS, Jung CK. CCND1 Splice Variant as A Novel Diagnostic and Predictive Biomarker for Thyroid Cancer. Cancers. 2018; 10(11):437. https://doi.org/10.3390/cancers10110437
Chicago/Turabian StyleJeon, Sora, Yourha Kim, Young Mun Jeong, Ja Seong Bae, and Chan Kwon Jung. 2018. "CCND1 Splice Variant as A Novel Diagnostic and Predictive Biomarker for Thyroid Cancer" Cancers 10, no. 11: 437. https://doi.org/10.3390/cancers10110437