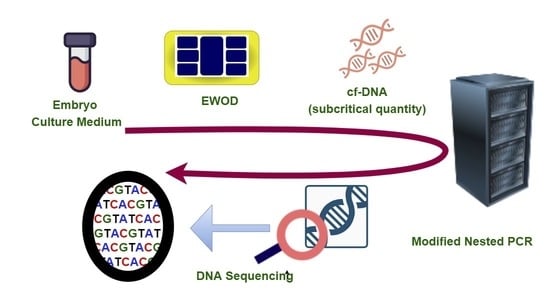
DNA Sequencing from Subcritical Concentration of Cell-Free DNA Extracted from Electrowetting-on-Dielectric Platform
Abstract
:1. Introduction
2. Materials and Methods
2.1. Calculation of Extraction Percentage by g-DNA
2.2. EWOD cf-DNA Extraction from Mouse Embryo Culture Medium
2.3. Amplification of Subcritical Concentration of EWOD Extracted cf-DNA
2.4. DNA Sequencing and BLAST Sequence Similarity Validation
3. Results
3.1. Recovery Rate of g-DNA in Conventional and EWOD Way
3.2. Extraction Quantity of cf-DNA after First and Second-Time q-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alias, A.B.; Chiang, C.-E.; Huang, H.-Y.; Lin, K.-T.; Lu, P.-J.; Wang, Y.-W.; Wu, T.-H.; Jiang, P.-S.; Chen, C.-A.; Yao, D.-J. Extraction of cell-free Dna from An embryo-culture Medium Using Micro-scale Bio-reagents on ewod. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Rojeab, A.Y. Magnetic properties govern the processes of DNA replication and the shortening of the telomere. Proc. World Acad. Sci. Eng. Technol. 2013, 8, 1342. [Google Scholar] [CrossRef]
- Shah, G.J.; Kim, C.-J.C. Meniscus-assisted high-efficiency magnetic collection and separation for EWOD droplet microfluidics. J. Microelectromech. Syst. 2009, 18, 363–375. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. Comparison of methods for the isolation of cell-free DNA from cell culture supernatant. Tumor Biol. 2020, 42, 1010428320916314. [Google Scholar] [CrossRef]
- Chiang, C.-E.; Huang, H.-Y.; Lin, K.-T.; Alias, A.B.; Lu, P.-J.; Wang, Y.-W.; Wu, T.-H.; Jiang, P.-S.; Chen, C.-A.; Yao, D.-J. A medical innovation: A new and improved method of DNA extraction with electrowetting-on-dielectric of genetic testing in-vitro fertilization (IVF). Microfluid. Nanofluidics 2020, 24, 1–9. [Google Scholar] [CrossRef]
- Malic, L.; Brassard, D.; Veres, T.; Tabrizian, M. Integration and detection of biochemical assays in digital microfluidic LOC devices. Lab Chip 2010, 10, 418–431. [Google Scholar] [CrossRef]
- Paik, P.; Pamula, V.K.; Pollack, M.G.; Fair, R.B. Electrowetting-based droplet mixers for microfluidic systems. Lab Chip 2003, 3, 28–33. [Google Scholar] [CrossRef]
- Wheeler, A.R.; Moon, H.; Kim, C.-J.C.; Loo, J.A.; Garrell, R.L. Electrowetting-based microfluidics for analysis of peptides and proteins by matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 2004, 76, 4833–4838. [Google Scholar] [CrossRef]
- Fobel, R.; Fobel, C.; Wheeler, A.R. DropBot: An open-source digital microfluidic control system with precise control of electrostatic driving force and instantaneous drop velocity measurement. Appl. Phys. Lett. 2013, 102, 193513. [Google Scholar] [CrossRef] [Green Version]
- Margulies, M.; Egholm, M.; Altman, W.E.; Attiya, S.; Bader, J.S.; Bemben, L.A.; Berka, J.; Braverman, M.S.; Chen, Y.-J.; Chen, Z. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005, 437, 376–380. [Google Scholar] [CrossRef]
- Kanagal-Shamanna, R.; Portier, B.P.; Singh, R.R.; Routbort, M.J.; Aldape, K.D.; Handal, B.A.; Rahimi, H.; Reddy, N.G.; Barkoh, B.A.; Mishra, B.M. Next-generation sequencing-based multi-gene mutation profiling of solid tumors using fine needle aspiration samples: Promises and challenges for routine clinical diagnostics. Mod. Pathol. 2014, 27, 314–327. [Google Scholar] [CrossRef] [Green Version]
- Kontchou, J.A.; Nocker, A. Optimization of viability qPCR for selective detection of membrane-intact Legionella pneumophila. J. Microbiol. Methods 2019, 156, 68–76. [Google Scholar] [CrossRef]
- Zachar, V.; Thomas, R.A.; Goustin, A.S. Absolute quantification of target DNA: A simple competitive PCR for efficient analysis of multiple samples. Nucleic Acids Res. 1993, 21, 2017. [Google Scholar] [CrossRef]
- Larionov, A.; Krause, A.; Miller, W. A standard curve based method for relative real time PCR data processing. BMC Bioinform. 2005, 6, 62. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Cai, K.; Zhang, R.; He, X.; Shen, X.; Liu, J.; Xu, J.; Qiu, F.; Lei, W.; Wang, J. Novel one-step single-tube nested quantitative real-time PCR assay for highly sensitive detection of SARS-CoV-2. Anal. Chem. 2020, 92, 9399–9404. [Google Scholar] [CrossRef]
- Tran, T.M.; Aghili, A.; Li, S.; Ongoiba, A.; Kayentao, K.; Doumbo, S.; Traore, B.; Crompton, P.D. A nested real-time PCR assay for the quantification of Plasmodium falciparum DNA extracted from dried blood spots. Malar. J. 2014, 13, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Neuberger, E.; Perez, I.; Le Guiner, C.; Moser, D.; Ehlert, T.; Allais, M.; Moullier, P.; Simon, P.; Snyder, R. Establishment of two quantitative nested qPCR assays targeting the human EPO transgene. Gene Ther. 2016, 23, 330–339. [Google Scholar] [CrossRef]
- Haff, L.A. Improved quantitative PCR using nested primers. Genome Res. 1994, 3, 332–337. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Ye, J.; McGinnis, S.; Madden, T.L. BLAST: Improvements for better sequence analysis. Nucleic Acids Res. 2006, 34, W6–W9. [Google Scholar] [CrossRef] [Green Version]
- Korf, I.; Yandell, M.; Bedell, J. Blast; O’Reilly Media, Inc.: Newton, MA, USA, 2003. [Google Scholar]
- Alias, A.B.; Huang, H.-Y.; Yao, D.-J. A Review on Microfluidics: An Aid to Assisted Reproductive Technology. Molecules 2021, 26, 4354. [Google Scholar] [CrossRef]





| Sample Type | DNA Sequence |
|---|---|
| Second-time PCR product of E2.5 with forward primer | TAAAAGTATAACCCCCTTGAGAATTAAAATGAACGAAAATCTATTTGCCTCATTCATTACCCCAAC |
| Second-time PCR product of E2.5 with reverse primer | GGGTTTTTACTTTATATGGTTGTTAGTGATTTTGGTGAAGGTGCCAGTGGGAATGTTTGTGATGAGACA |
| Second-time PCR product of E3.5 with forward primer | TAAAAGTATAACCCCCTATGAGAATTAAAATGAACGAAAATCTATTTGCCTCATTCATTACCCCAAC |
| Second-time PCR product of E3.5 with reverse primer | GGGTTTTTACTTTATATGGTTGTTAGTGATTTTGGTGAAGGTGCCAGTGGGAATGTTTGTGATGAGACA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alias, A.B.; Huang, H.-Y.; Wang, Y.-W.; Lin, K.-T.; Lu, P.-J.; Wu, T.-H.; Jiang, P.-S.; Chen, C.-A.; Yao, D.-J. DNA Sequencing from Subcritical Concentration of Cell-Free DNA Extracted from Electrowetting-on-Dielectric Platform. Micromachines 2022, 13, 507. https://doi.org/10.3390/mi13040507
Alias AB, Huang H-Y, Wang Y-W, Lin K-T, Lu P-J, Wu T-H, Jiang P-S, Chen C-A, Yao D-J. DNA Sequencing from Subcritical Concentration of Cell-Free DNA Extracted from Electrowetting-on-Dielectric Platform. Micromachines. 2022; 13(4):507. https://doi.org/10.3390/mi13040507
Chicago/Turabian StyleAlias, Anand Baby, Hong-Yuan Huang, Yi-Wen Wang, Kai-Ti Lin, Pei-Jhen Lu, Tzu-Hui Wu, Pei-Shin Jiang, Chien-An Chen, and Da-Jeng Yao. 2022. "DNA Sequencing from Subcritical Concentration of Cell-Free DNA Extracted from Electrowetting-on-Dielectric Platform" Micromachines 13, no. 4: 507. https://doi.org/10.3390/mi13040507