Micromirror-Embedded Coverslip Assembly for Bidirectional Microscopic Imaging
Abstract
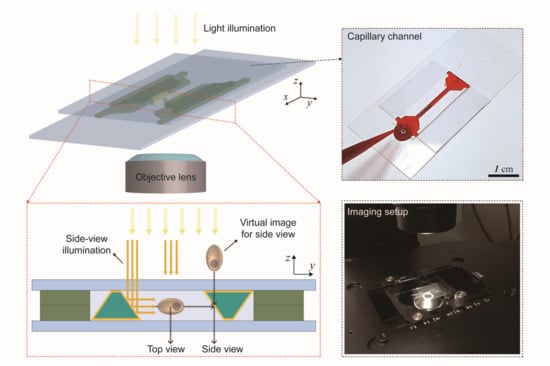
:1. Introduction
2. Materials and Methods
2.1. Preparation of a PDMS Channel for Micromirror Capillary Molding
2.2. Fabrication of Coverslip with Parallel Column Structures
2.3. Preparation of Beads and Cells
3. Results and Discussion
3.1. Fabrication of Trapezoidal Micromirror and Micromirror-Embedded Coverslip Assembly
3.2. Bidirectional Imaging of Fluorescent Beads with the Micromirror-Embedded Coverslip
3.3. Bidirectional Imaging of Live Cells with the Micromirror-Embedded Coverslip
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lichtman, J.W.; Conchello, J.-A. Fluorescence microscopy. Nat. Methods 2005, 2, 910–919. [Google Scholar] [CrossRef] [PubMed]
- Leung, B.O.; Chou, K.C. Review of super-resolution fluorescence microscopy for biology. Appl. Spectrosc. 2011, 65, 967–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizutani, R.; Suzuki, Y. X-ray microtomography in biology. Micron 2012, 43, 104–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jo, Y.; Cho, H.; Lee, S.Y.; Choi, G.; Kim, G.; Min, H.-S.; Park, Y. Quantitative phase imaging and artificial intelligence: A review. IEEE J. Sel. Top. Quantum Electron. 2018, 25, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Popescu, G.; Park, Y.; Lue, N.; Best-Popescu, C.; Deflores, L.; Dasari, R.R.; Feld, M.S.; Badizadegan, K. Optical imaging of cell mass and growth dynamics. Am. J. Physiol. Cell Physiol. 2008, 295, C538–C544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Song, C.; Chen, C.-C.; Xu, R.; Raines, K.S.; Fahimian, B.P.; Lu, C.-H.; Lee, T.-K.; Nakashima, A.; Urano, J. Quantitative 3D imaging of whole, unstained cells by using X-ray diffraction microscopy. Proc. Natl. Acad. Sci. USA 2010, 107, 11234–11239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heymann, J.A.; Hayles, M.; Gestmann, I.; Giannuzzi, L.A.; Lich, B.; Subramaniam, S. Site-specific 3D imaging of cells and tissues with a dual beam microscope. J. Struct. Biol. 2006, 155, 63–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tata, B.; Raj, B. Confocal laser scanning microscopy: Applications in material science and technology. Bull. Mater. Sci. 1998, 21, 263–278. [Google Scholar] [CrossRef] [Green Version]
- Patel, D.V.; McGhee, C.N. Quantitative analysis of in vivo confocal microscopy images: A review. Surv. Ophthalmol. 2013, 58, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Paddock, S.W. Principles and practices of laser scanning confocal microscopy. Mol. Biotechnol. 2000, 16, 127–149. [Google Scholar] [CrossRef]
- Lidke, D.S.; Lidke, K.A. Advances in high-resolution imaging–techniques for three-dimensional imaging of cellular structures. J. Cell Sci. 2012, 125, 2571–2580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sung, Y.; Choi, W.; Fang-Yen, C.; Badizadegan, K.; Dasari, R.R.; Feld, M.S. Optical diffraction tomography for high resolution live cell imaging. Opt. Express 2009, 17, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Wedberg, T.C.; Stamnes, J.J. Quantitative imaging by optical diffraction tomography. Opt. Rev. 1995, 2, 28–31. [Google Scholar] [CrossRef]
- Choi, W.; Fang-Yen, C.; Badizadegan, K.; Oh, S.; Lue, N.; Dasari, R.R.; Feld, M.S. Tomographic phase microscopy. Nat. Methods 2007, 4, 717–719. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Kim, K.S.; Park, H.; Ye, J.C.; Park, Y. Real-time visualization of 3-D dynamic microscopic objects using optical diffraction tomography. Opt. Express 2013, 21, 32269–32278. [Google Scholar] [CrossRef] [PubMed]
- Charrière, F.; Marian, A.; Montfort, F.; Kuehn, J.; Colomb, T.; Cuche, E.; Marquet, P.; Depeursinge, C. Cell refractive index tomography by digital holographic microscopy. Opt. Lett. 2006, 31, 178–180. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Shim, H.; Kim, K.; Park, H.; Jang, S.; Park, Y. Profiling individual human red blood cells using common-path diffraction optical tomography. Sci. Rep. 2014, 4, 6659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, J.; Kim, K.; Park, H.; Choi, C.; Jang, S.; Park, Y. Label-free characterization of white blood cells by measuring 3D refractive index maps. Biomed. Opt. Express 2015, 6, 3865–3875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merola, F.; Memmolo, P.; Miccio, L.; Savoia, R.; Mugnano, M.; Fontana, A.; D’ippolito, G.; Sardo, A.; Iolascon, A.; Gambale, A. Tomographic flow cytometry by digital holography. Light Sci. Appl. 2017, 6, e16241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, S.; Kim, J.; Lee, J.-R.; Jeon, E.-c.; Je, T.-J.; Lee, W.; Park, Y. Enhancement of optical resolution in three-dimensional refractive-index tomograms of biological samples by employing micromirror-embedded coverslips. Lab. A Chip 2018, 18, 3484–3491. [Google Scholar] [CrossRef]
- Bates, K.E.; Lu, H. Optics-integrated microfluidic platforms for biomolecular analyses. Biophys. J. 2016, 110, 1684–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Zhang, Y.; Chen, S.; Hao, R. Micro-optical Components for Bioimaging on Tissues, Cells and Subcellular Structures. Micromachines 2019, 10, 405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Justice, B.A.; Badr, N.A.; Felder, R.A. 3D cell culture opens new dimensions in cell-based assays. Drug Discov. Today 2009, 14, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Tibbitt, M.W.; Anseth, K.S. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 2009, 103, 655–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravi, M.; Paramesh, V.; Kaviya, S.; Anuradha, E.; Solomon, F.P. 3D cell culture systems: Advantages and applications. J. Cell. Physiol. 2015, 230, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Merlos, A.; Acero, M.; Bao, M.; Bausells, J.; Esteve, J. TMAH/IPA anisotropic etching characteristics. Sens. Actuators A Phys. 1993, 37, 737–743. [Google Scholar] [CrossRef]
- Powell, O.; Harrison, H.B. Anisotropic etching of {100} and {110} planes in (100) silicon. J. Micromech. Microeng. 2001, 11, 217. [Google Scholar] [CrossRef]
- Koh, J.; Kim, J.; Shin, J.H.; Lee, W. Fabrication and integration of microprism mirrors for high-speed three-dimensional measurement in inertial microfluidic system. Appl. Phys. Lett. 2014, 105, 114103. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-a.; Lee, J.-R.; Je, T.-J.; Jeon, E.-c.; Lee, W. Size-dependent inertial focusing position shift and particle separations in triangular microchannels. Anal. Chem. 2018, 90, 1827–1835. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-a.; Kommajosula, A.; Choi, Y.-h.; Lee, J.-R.; Jeon, E.-C.; Ganapathysubramanian, B.; Lee, W. Inertial focusing in triangular microchannels with various apex angles. Biomicrofluidics 2020, 14, 024105. [Google Scholar] [CrossRef] [PubMed]
- Karp, J.H.; Tremblay, E.J.; Ford, J.E. Planar micro-optic solar concentrator. Opt. Express 2010, 18, 1122–1133. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.; Jeong, S.; Choi, H.J.; Shin, N.; Kim, B.; Jeon, E.c.; Lee, J.Y. Toward perfect light trapping in thin-film photovoltaic cells: Full utilization of the dual characteristics of light. Adv. Opt. Mater. 2015, 3, 1697–1702. [Google Scholar] [CrossRef]
- Lee, J.-R.; Je, T.-J.; Woo, S.; Yoo, Y.-E.; Jeong, J.-H.; Jeon, E.-C.; Kim, H. Geometric and Wave Optic Features in the Optical Transmission Patterns of Injection-molded Mesoscale Pyramid Prism Patterned Plates. Curr. Opt. Photonics 2018, 2, 140–146. [Google Scholar]
- Nam, S.M.; Kim, K.; Kang, I.-S.; Park, W.; Lee, W. Generation of 3D Microparticles in Microchannels with Non-rectangular Cross Sections. Biochip J. 2019, 13, 226–235. [Google Scholar]
- O’Brien, A.K.; Bowman, C.N. Impact of oxygen on photopolymerization kinetics and polymer structure. Macromolecules 2006, 39, 2501–2506. [Google Scholar]






| Intensity Ratio | Roundness | Aspect Ratio | Distance from the Mirror (μm) | ||||
|---|---|---|---|---|---|---|---|
| Horizontal | Vertical | Top | Side | Top | Side | ||
| B1 | 0.97 | 0.963 | 0.998 | 0.971 | 1.002 | 1.03 | 43.4 |
| B2 | 0.978 | 0.978 | 0.998 | 0.988 | 1.002 | 1.012 | 17.1 |
| B3 | 0.957 | 0.957 | 0.994 | 0.951 | 1.006 | 1.052 | 97.5 |
| B4 | 0.972 | 0.979 | 0.994 | 0.981 | 1.006 | 1.019 | 20.7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, D.; Kim, J.; Song, E.; Jeong, J.-Y.; Jeon, E.-c.; Kim, P.; Lee, W. Micromirror-Embedded Coverslip Assembly for Bidirectional Microscopic Imaging. Micromachines 2020, 11, 582. https://doi.org/10.3390/mi11060582
Lee D, Kim J, Song E, Jeong J-Y, Jeon E-c, Kim P, Lee W. Micromirror-Embedded Coverslip Assembly for Bidirectional Microscopic Imaging. Micromachines. 2020; 11(6):582. https://doi.org/10.3390/mi11060582
Chicago/Turabian StyleLee, Dongwoo, Jihye Kim, Eunjoo Song, Ji-Young Jeong, Eun-chae Jeon, Pilhan Kim, and Wonhee Lee. 2020. "Micromirror-Embedded Coverslip Assembly for Bidirectional Microscopic Imaging" Micromachines 11, no. 6: 582. https://doi.org/10.3390/mi11060582