Hyperspectral Image Classification via Spatial Shuffle-Based Convolutional Neural Network
Abstract
:1. Introduction
2. Proposed Method
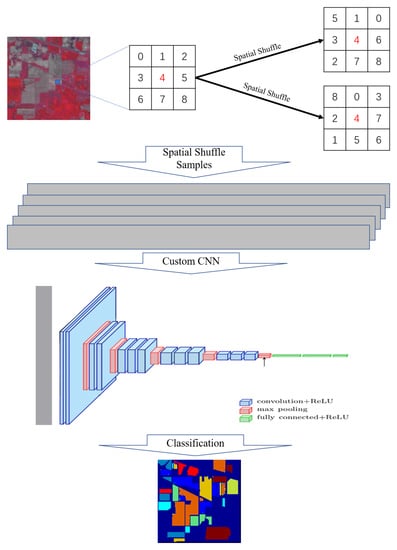
2.1. Spatial Shuffle
2.2. Basic CNN
3. Experiments and Results
3.1. Dataset
3.2. Parameter Settings
3.3. Results on the IP Dataset
3.4. Results on the SV Dataset
3.5. Results on the UP Dataset
4. Discussion
4.1. Classification Ability with Less Samples
4.2. Effects of Neighborhood Sizes
4.3. Effects of Spatial Shuffle
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gwon, Y.; Kim, D.; You, H.J.; Nam, S.H.; Kim, Y.D. A Standardized Procedure to Build a Spectral Library for Hazardous Chemicals Mixed in River Flow Using Hyperspectral Image. Remote Sens. 2023, 15, 477. [Google Scholar] [CrossRef]
- Shitharth, S.; Manoharan, H.; Alshareef, A.M.; Yafoz, A.; Alkhiri, H.; Mirza, O.M. Hyper spectral image classifications for monitoring harvests in agriculture using fly optimization algorithm. Comput. Electr. Eng. 2022, 103, 108400. [Google Scholar]
- Verma, R.K.; Sharma, L.K.; Lele, N. AVIRIS-NG hyperspectral data for biomass modeling: From ground plot selection to forest species recognition. J. Appl. Remote Sens. 2023, 17, 014522. [Google Scholar]
- Yang, H.Q.; Chen, C.W.; Ni, J.H.; Karekal, S. A hyperspectral evaluation approach for quantifying salt-induced weathering of sandstone. Sci. Total Environ. 2023, 885, 163886. [Google Scholar] [CrossRef]
- Calin, M.A.; Calin, A.C.; Nicolae, D.N. Application of airborne and spaceborne hyperspectral imaging techniques for atmospheric research: Past, present, and future. Appl. Spectrosc. Rev. 2021, 56, 289–323. [Google Scholar]
- Cui, J.; Yan, B.K.; Wang, R.S.; Tian, F.; Zhao, Y.J.; Liu, D.C.; Yang, S.M.; Shen, W. Regional-scale mineral mapping using ASTER VNIR/SWIR data and validation of reflectance and mineral map products using airborne hyperspectral CASI/SASI data. Int. J. Appl. Earth Obs. Geoinf. 2014, 33, 127–141. [Google Scholar]
- Kumar, V.; Ghosh, J. Camouflage detection using MWIR hyperspectral images. J. Indian Soc. Remote Sens. 2017, 45, 139–145. [Google Scholar] [CrossRef]
- Shimoni, M.; Haelterman, R.; Perneel, C. Hypersectral imaging for military and security applications: Combining myriad processing and sensing techniques. IEEE Geosci. Remote Sens. Mag. 2019, 7, 101–117. [Google Scholar] [CrossRef]
- Liu, J.J.; Wu, Z.B.; Li JPlaza, A.; Yuan, Y.H. Probabilistic-kernel collaborative representation for spatial–spectral hyper-spectral image classification. IEEE Trans. Geosci. Remote Sens. 2015, 54, 2371–2384. [Google Scholar] [CrossRef]
- Wu, L.; Huang, J.; Guo, M.S. Multidimensional Low-Rank Representation for Sparse Hyperspectral Unmixing. IEEE Geosci. Remote Sens. Lett. 2023, 20, 5502805. [Google Scholar] [CrossRef]
- Plaza, A.; Benediktsson, J.A.; Boardman, J.W.; Brazile, B.; Bruzzone, L.; Camps-Valls, G.; Chanussot, J.; Fauvel, M.; Gamba, P.; Gualtieri, A.; et al. Recent advances in techniques for hyperspectral image processing. Remote Sens. Environ. 2009, 113, S110–S122. [Google Scholar] [CrossRef]
- Li, J.; Marpu, P.R.; Plaza, A.; GenBioucas-Dias, J.M.; Benediktsson, J.A. Geralized composite kernel framework for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2013, 51, 4816–4829. [Google Scholar] [CrossRef]
- Zhang, Y.Q.; Cao, G.; Li, X.S.; Wang, B.S. Cascaded random forest for hyperspectral image classification. IEEE J. Sel. Top. Appl. Earth Observ. Remote Sens. 2018, 11, 1082–1094. [Google Scholar] [CrossRef]
- Gao, B.T.; Yu, L.F.; Ren, L.L.; Zhan, Z.Y.; Luo, Y.Q. Early Detection of Dendroctonus valens Infestation at Tree Level with a Hyperspectral UAV Image. Remote Sens. 2023, 15, 407. [Google Scholar] [CrossRef]
- Xia, J.S.; Du, P.J.; He, X.Y.; Chanussot, J. Hyperspectral remote sensing image classification based on rotation forest. IEEE Geosci. Remote Sens. Lett. 2013, 11, 239–243. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Huang, Y.Y.; Wei, L.; Zhang, F.; Li, H.C. Deep convolutional neural networks for hyperspectral image classification. J. Sens. 2015, 2015, 258619. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.F.; Ye, Y.M.; Li, X.T.; Lau, R.Y.K.; Zhang, X.F.; Huang, X.H. Hyperspectral image classification with deep learning models. IEEE Trans. Geosci. Remote Sens. 2018, 56, 5408–5423. [Google Scholar] [CrossRef]
- Ma, X.T.; Man, Q.X.; Yang, X.M.; Dong, P.L.; Yang, Z.L.; Wu, J.R.; Liu, C.H. Urban Feature Extraction within a Complex Urban Area with an Improved 3D-CNN Using Airborne Hyperspectral Data. Remote Sens. 2023, 15, 992. [Google Scholar] [CrossRef]
- Mou, L.; Ghamisi, P.; Zhu, X.X. Deep recurrent neural networks for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 3639–3655. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.K.; Liu, B.; He, P.P.; Hu, Q.F.; Gao, K.L.; Li, H. Masked Graph Convolutional Network for Small Sample Classification of Hyperspectral Images. Remote Sens. 2023, 15, 1869. [Google Scholar] [CrossRef]
- Chen, Y.S.; Lin, Z.H.; Zhao, X.; Wang, G.; Gu, Y.F. Deep learning-based classification of hyperspectral data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2094–2107. [Google Scholar] [CrossRef]
- Zhu, L.; Chen, Y.S.; Ghamisi, P.; Benediktsson, J.A. Generative adversarial networks for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2018, 56, 5046–5063. [Google Scholar] [CrossRef]
- Paoletti, M.E.; Haut, J.M.; Fernandez-Beltran, R.; Plaza, J.; Plaza, A.; Li, J.; Pla, F. Capsule networks for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2018, 57, 2145–2160. [Google Scholar] [CrossRef]
- Hang, R.L.; Liu, Q.S.; Hong, D.F.; Ghamisi, P. Cascaded recurrent neural networks for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2019, 57, 5384–5394. [Google Scholar] [CrossRef] [Green Version]
- Hong, D.F.; Gao, L.R.; Yao, J.; Zhang, B.; Plaza, A.; Chanussot, J. Graph convolutional networks for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2021, 59, 5966–5978. [Google Scholar] [CrossRef]
- Hong, D.; Han, Z.; Yao, J.; Gao, L.R.; Zhang, B.; Plaza, A.; Chanussot, J. SpectralFormer: Rethinking Hyperspectral Image Classification With Transformers. IEEE Trans. Geosci. Remote Sens. 2022, 60, 5518615. [Google Scholar] [CrossRef]
- Wu, H.; Prasad, S. Semi-supervised deep learning using pseudo labels for hyperspectral image classification. IEEE Trans. Image Process. 2018, 27, 1259–1270. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.X.; Ge, L.Y.; Chen, G.; Radenkovic, M.; Wang, X.P.; Duan, J.M.; Pan, Z.K. Nonlocal graph theory based transductive learning for hyperspectral image classification. Pattern Recognit. 2021, 116, 107967. [Google Scholar] [CrossRef]
- Li, J.; Bioucas-dias, J.M.; Plaza, A. Semisupervised hyperspectral image classification using soft sparse multinomial logistic regression. IEEE Geosci. Remote Sens. Lett. 2013, 10, 318–322. [Google Scholar]
- Fang, B.; Li, Y.; Zhang, H.K.; Chan, J.C.W. Collaborative learning of lightweight convolutional neural network and deep clustering for hyperspectral image semi-supervised classification with limited training samples. ISPRS J. Photogramm. Remote Sens. 2020, 161, 164–178. [Google Scholar]
- Zhang, C.; Yue, J.; Qin, Q. Global prototypical network for few-shot hyperspectral image classification. IEEE J. Sel. Top. Appl. Earth Observ. Remote Sens. 2020, 13, 4748–4759. [Google Scholar] [CrossRef]
- Gao, K.L.; Liu, B.; Yu, X.C.; Qin, J.C.; Zhang, P.Q.; Tan, X. Deep relation network for hyperspectral image few-shot classification. Remote Sens. 2020, 12, 923. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.K.; Liu, M.; Chen, Y.S.; Xu, Y.M.; Li, W.; Du, Q. Deep cross-domain few-shot learning for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2022, 60, 1–18. [Google Scholar] [CrossRef]
- Paoletti, M.E.; Haut, J.M.; Plaza, J.; Plaza, A. Deep learning classifers for hyperspectral imaging: A review. ISPRS J. Photogramm. Remote Sens. 2019, 158, 279–317. [Google Scholar] [CrossRef]
- Ghamisi, P.; Maggiori, E.; Li, S.T.; Souza, R.; Tarablaka, Y.; Moser, G.; De Giorgi, A.; Fang, L.Y.; Chen, Y.S.; Chi, M.M.; et al. New frontiers in spectral-spatial hyperspectral image classifcation: The latest advances based on mathematical morphology, markov random felds, segmentation, sparse representation, and deep learning. IEEE Geosci. Remote Sens. Mag. 2018, 6, 10–43. [Google Scholar]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; pp. 3320–3328. [Google Scholar]
- Li, W.; Wu, G.D.; Zhang, F.; Du, Q. Hyperspectral Image Classification Using Deep Pixel-Pair Features. IEEE Trans. Geosci. Remote Sens. 2017, 55, 844–853. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef] [Green Version]
- Haut, J.; Paoletti, M.; Paz-Gallardo, A.; Plaza, J.; Plaza, A. Cloud implementation of logistic regression for hyperspectral image classifcation. In Proceedings of the 17th International Conference on Computational and Mathematical Methods in Science and Engineering, CMMSE, Rota, Spain, 4–8 July 2017; Vigo-Aguiar, J., Ed.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1063–2321. [Google Scholar]
- Li, J.; Zhao, X.; Li, Y.; Du, Q.; Xi, B.; Hu, J. Classifcation of hyperspectral imagery using a new fully convolutional neural network. IEEE Geosci. Remote Sens. Lett. 2018, 15, 292–296. [Google Scholar] [CrossRef]
- Ham, J.; Chen, Y.; Crawford, M.M.; Ghosh, J. Investigation of the random forest framework for classifcation of hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2005, 43, 492–501. [Google Scholar] [CrossRef] [Green Version]











| IP | SV | UP | |
|---|---|---|---|
| Seq_1 | Conv-BN-ReLU, (1 × 3), 32 | Conv-BN-ReLU, (1 × 3), 32 | Conv-BN-ReLU, (1 × 3), 32 |
| Conv-BN-ReLU, (1 × 3), 32 | Conv-BN-ReLU, (1 × 3), 32 | Conv-BN-ReLU, (1 × 3), 32 | |
| Conv-BN-ReLU, (1 × 3), 32 | Conv-BN-ReLU, (1 × 3), 32 | Conv-BN-ReLU, (1 × 3), 32 | |
| Conv-BN-ReLU, (1 × 3), 32 | Conv-BN-ReLU, (1 × 3), 32 | Conv-BN-ReLU, (1 × 3), 32 | |
| Conv-BN-ReLU, (1 × 3), 32 | Conv-BN-ReLU, (1 × 3), 32 | Conv-BN-ReLU, (1 × 3), 32 | |
| Conv-BN-ReLU, (1 × 3), 32 | Conv-BN-ReLU, (1 × 3), 32 | Conv-BN-ReLU, (1 × 3), 32 | |
| Maxpool_2 × 1 | Maxpool_2 × 1 | Maxpool_2 × 1 | |
| Conv-BN-ReLU, (3 × 1), 32 | Conv-BN-ReLU, (3 × 1), 32 | Conv-BN-ReLU, (3 × 1), 32 | |
| Conv-BN-ReLU, (3 × 1), 32 | Conv-BN-ReLU, (3 × 1), 32 | Conv-BN-ReLU, (3 × 1), 32 | |
| Maxpool_2 × 1 | Maxpool_2 × 1 | Maxpool_2 × 1 | |
| Conv-BN-ReLU, (3 × 1), 32 | Conv-BN-ReLU, (3 × 1), 32 | Conv-BN-ReLU, (3 × 1), 32 | |
| Maxpool_1 × 2 | Maxpool_1 × 2 | Maxpool_1 × 2 | |
| Seq_2 | Conv-BN-ReLU, (1 × 3), 32 | Conv-BN-ReLU, (1 × 3), 32 | Conv-BN-ReLU, (1 × 3), 32 |
| Conv-BN-ReLU, (1 × 3), 32 | Conv-BN-ReLU, (1 × 3), 32 | Conv-BN-ReLU, (1 × 3), 32 | |
| Conv-BN-ReLU, (1 × 3), 32 | Conv-BN-ReLU, (1 × 3), 32 | Conv-BN-ReLU, (1 × 3), 32 | |
| Maxpool_1 × 2 | Maxpool_1 × 2 | Maxpool_1 × 2 | |
| Seq_3 | Conv-BN-ReLU, (1 × 3), 64 | Conv-BN-ReLU, (1 × 3), 64 | Conv-BN-ReLU, (1 × 3), 64 |
| Conv-BN-ReLU, (1 × 3), 64 | Conv-BN-ReLU, (1 × 3), 64 | Conv-BN-ReLU, (1 × 3), 64 | |
| Conv-BN-ReLU, (1 × 3), 64 | Conv-BN-ReLU, (1 × 3), 64 | Maxpool_1 × 2 | |
| Maxpool_1 × 2 | Maxpool_1 × 2 | ||
| Seq_4 | Conv-BN-ReLU, (1 × 3), 64 | Conv-BN-ReLU, (1 × 3), 64 | Conv-BN-ReLU, (1 × 3), 64 |
| Conv-BN-ReLU, (1 × 3), 64 | Conv-BN-ReLU, (1 × 3), 64 | Conv-BN-ReLU, (1 × 3), 64 | |
| Conv-BN-ReLU, (1 × 3), 64 | Conv-BN-ReLU, (1 × 3), 64 | Conv-BN-ReLU, (1 × 3), 64 | |
| Maxpool_1 × 2 | Maxpool_1 × 2 | ||
| Seq_5 | Conv-BN-ReLU, (1 × 3), 64 | Conv-BN-ReLU, (1 × 3), 64 | |
| Conv-BN-ReLU, (1 × 3), 64 | Conv-BN-ReLU, (1 × 3), 64 | ||
| Conv-BN-ReLU, (1 × 3), 64 | |||
| Seq_6 | FC-64 | FC-64 | FC-64 |
| FC-classnum | FC-classnum | FC-classnum |
| No. | Class Name | Training Num | Testing Num | All Num |
|---|---|---|---|---|
| 0 | Background | - | - | 10,776 |
| 1 | Alfalfa | - | - | 46 |
| 2 | Corn-notill | 200 | 1228 | 1428 |
| 3 | Corn-min | 200 | 630 | 830 |
| 4 | Corn | - | - | 237 |
| 5 | Grass/Pasture | 200 | 283 | 483 |
| 6 | Grass/Trees | 200 | 530 | 730 |
| 7 | Grass/pasture-mowed | - | - | 28 |
| 8 | Hay-windrowed | 200 | 278 | 478 |
| 9 | Oats | - | - | 20 |
| 10 | Soybeans-notill | 200 | 772 | 972 |
| 11 | Soybeans-min | 200 | 2255 | 2455 |
| 12 | Soybean-clean | 200 | 393 | 593 |
| 13 | Wheat | - | - | 205 |
| 14 | Woods | 200 | 1065 | 1265 |
| 15 | Bldg-Grass-Tree-Drives | - | - | 386 |
| 16 | Stone-steel towers | - | - | 93 |
| Total | 1800 | 7434 | 21,025 |
| No. | Class Name | Training Num | Testing Num | All Num |
|---|---|---|---|---|
| 0 | Background | - | - | 56,975 |
| 1 | Brocoli-gree-weeds-1 | 200 | 1809 | 2009 |
| 2 | Brocoli-gree-weeds-2 | 200 | 3526 | 3726 |
| 3 | Fallow | 200 | 1776 | 1976 |
| 4 | Fallow-rough-plow | 200 | 1194 | 1394 |
| 5 | Fallow-smooth | 200 | 2478 | 2678 |
| 6 | Stubble | 200 | 3759 | 3959 |
| 7 | Celery | 200 | 3379 | 3579 |
| 8 | Grapes-untrained | 200 | 11,071 | 11,271 |
| 9 | Soil-vinyard-develop | 200 | 6003 | 6203 |
| 10 | Corn-senesced-green-weeds | 200 | 3078 | 3278 |
| 11 | Lettuce-romaine-4wk | 200 | 868 | 1068 |
| 12 | Lettuce-romaine-5wk | 200 | 1727 | 1927 |
| 13 | Lettuce-romaine-6wk | 200 | 716 | 916 |
| 14 | Lettuce-romaine-7wk | 200 | 870 | 1070 |
| 15 | Vinyard-untrained | 200 | 7068 | 7268 |
| 16 | Vinyard-vertical-trellis | 200 | 1607 | 1807 |
| Total | 3200 | 50,929 | 111,104 |
| No. | Class Name | Training Num | Testing Num | All Num |
|---|---|---|---|---|
| 0 | Background | - | - | 164,624 |
| 1 | Asphalt | 200 | 6431 | 6631 |
| 2 | Meadows | 200 | 18,449 | 18,649 |
| 3 | Gravel | 200 | 1899 | 2099 |
| 4 | Trees | 200 | 2864 | 3064 |
| 5 | Painted metal sheets | 200 | 1145 | 1345 |
| 6 | Bare Soil | 200 | 4829 | 5029 |
| 7 | Bitumen | 200 | 1130 | 1330 |
| 8 | Self-Blocking Bricks | 200 | 3482 | 3682 |
| 9 | Shadows | 200 | 747 | 947 |
| Total | 1800 | 40,976 | 207,400 |
| MLR | SVM | RF | ELM | CNN2D | PPF | Proposed | |
|---|---|---|---|---|---|---|---|
| Corn-notill | 42.59 | 63.84 | 61.97 | 78.34 | 88.03 | 97.31 | 98.53 |
| Corn-min | 37.44 | 66.20 | 68.63 | 81.98 | 96.19 | 95.49 | 99.65 |
| Grass/Pasture | 70.52 | 95.52 | 92.16 | 95.15 | 98.13 | 99.25 | 99.25 |
| Grass/Trees | 95.28 | 100.00 | 98.11 | 100.00 | 100.00 | 99.81 | 100.00 |
| Hay-windrowed | 100.00 | 100.00 | 99.64 | 100.00 | 100.00 | 100.00 | 100.00 |
| Soybeans-notill | 62.58 | 72.10 | 84.22 | 85.01 | 95.31 | 95.70 | 97.52 |
| Soybeans-min | 62.00 | 63.76 | 64.35 | 61.68 | 76.23 | 86.94 | 96.25 |
| Soybean-clean | 36.64 | 84.48 | 79.39 | 93.64 | 99.75 | 100.00 | 100.00 |
| Woods | 87.79 | 98.12 | 96.62 | 98.40 | 100.00 | 99.81 | 99.72 |
| OA | 63.42 | 76.12 | 76.68 | 81.04 | 89.93 | 94.73 | 98.26 |
| AA | 66.09 | 82.67 | 82.79 | 88.24 | 94.85 | 97.15 | 98.99 |
| Kappa | 0.5647 | 0.7184 | 0.7259 | 0.7776 | 0.8810 | 0.9371 | 0.9792 |
| MLR | SVM | RF | ELM | CNN2D | PPF | Proposed | |
|---|---|---|---|---|---|---|---|
| Brocoli-gree-weeds-1 | 97.82 | 98.85 | 99.43 | 99.83 | 62.41 | 100 | 100 |
| Brocoli-gree-weeds-2 | 97.82 | 99.89 | 99.74 | 99.83 | 100 | 100 | 100 |
| Fallow | 92.06 | 99.38 | 99.04 | 97.75 | 99.94 | 99.77 | 99.94 |
| Fallow-rough-plow | 99.08 | 99.5 | 99.41 | 99.16 | 100 | 99.66 | 99.92 |
| Fallow-smooth | 97.7 | 98.14 | 97.54 | 98.71 | 97.58 | 98.35 | 99.56 |
| Stubble | 99.46 | 99.92 | 99.81 | 99.87 | 100 | 100 | 99.97 |
| Celery | 99.4 | 99.91 | 99.29 | 99.76 | 99.76 | 99.97 | 99.97 |
| Grapes-untrained | 70.44 | 85.03 | 61.66 | 83.83 | 89.59 | 83.92 | 95.71 |
| Soil-vinyard-develop | 96.21 | 99.48 | 98.83 | 99.92 | 99.92 | 99.9 | 99.98 |
| Corn-senesced-green-weeds | 85.44 | 94.37 | 88.28 | 94.37 | 94.64 | 98.51 | 98.84 |
| Lettuce-romaine-4wk | 92.44 | 97.52 | 93.15 | 94.92 | 99.41 | 100 | 99.65 |
| Lettuce-romaine-5wk | 99.64 | 99.82 | 97.63 | 99.23 | 99.94 | 100 | 100 |
| Lettuce-romaine-6wk | 98.86 | 99.71 | 98.15 | 99 | 100 | 99.57 | 100 |
| Lettuce-romaine-7wk | 91.19 | 98.24 | 94.83 | 94.36 | 99.65 | 99.29 | 99.18 |
| Vinyard-untrained | 62.78 | 69.36 | 69.64 | 69.41 | 73.93 | 85.82 | 94.33 |
| Vinyard-vertical-trellis | 91.08 | 98.76 | 98.27 | 98.69 | 99.72 | 99.45 | 99.93 |
| OA | 85.84 | 91.84 | 86 | 91.47 | 92.36 | 94.31 | 98.16 |
| AA | 91.96 | 96.12 | 93.42 | 95.54 | 94.78 | 97.76 | 99.19 |
| Kappa | 0.8417 | 0.9086 | 0.8441 | 0.9044 | 0.9142 | 0.9364 | 0.9794 |
| MLR | SVM | RF | ELM | CNN2D | PPF | Proposed | |
|---|---|---|---|---|---|---|---|
| Asphalt | 73.55 | 88.94 | 81.32 | 62.08 | 96.01 | 98.2 | 99.64 |
| Meadows | 75.77 | 93.72 | 78.4 | 91.2 | 90.46 | 97.78 | 99.53 |
| Gravel | 76.95 | 85.23 | 76.89 | 81.66 | 95.73 | 91.67 | 97.78 |
| Trees | 93.06 | 96.02 | 94.96 | 95.14 | 97.57 | 96.55 | 97.75 |
| Painted metal sheets | 99.21 | 99.65 | 99.56 | 99.48 | 100 | 99.91 | 100 |
| Bare Soil | 73.49 | 90.43 | 82.61 | 83.35 | 98.92 | 97.54 | 99.88 |
| Bitumen | 89.12 | 91.59 | 90.35 | 91.95 | 98.76 | 94.42 | 98.32 |
| Self-Blocking Bricks | 74.73 | 83.46 | 78.89 | 68.12 | 90.09 | 92.19 | 98.71 |
| Shadows | 99.87 | 100 | 100 | 99.87 | 100 | 99.87 | 100 |
| OA | 77.83 | 91.68 | 81.86 | 83.91 | 93.76 | 96.97 | 99.3 |
| AA | 83.97 | 92.12 | 87 | 85.87 | 96.39 | 96.46 | 99.07 |
| Kappa | 0.7152 | 0.8898 | 0.7662 | 0.7888 | 0.9181 | 0.9595 | 0.9906 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Cao, B.; Liu, J. Hyperspectral Image Classification via Spatial Shuffle-Based Convolutional Neural Network. Remote Sens. 2023, 15, 3960. https://doi.org/10.3390/rs15163960
Wang Z, Cao B, Liu J. Hyperspectral Image Classification via Spatial Shuffle-Based Convolutional Neural Network. Remote Sensing. 2023; 15(16):3960. https://doi.org/10.3390/rs15163960
Chicago/Turabian StyleWang, Zhihui, Baisong Cao, and Jun Liu. 2023. "Hyperspectral Image Classification via Spatial Shuffle-Based Convolutional Neural Network" Remote Sensing 15, no. 16: 3960. https://doi.org/10.3390/rs15163960