1. Introduction
The characteristics of individual plants or groups of plants can be used to evaluate crop growth [
1] and can reveal various levels of crop growth within regions [
2]. However, in precision agriculture, effective monitoring of growth conditions can provide not only real-time information for field management but also a basis for estimating crop yield [
2,
3,
4]. Traditionally, field managers determine crop-growth status by visual inspection, which is laborious and time-consuming. However, in precision agriculture, remote sensing technology has become increasingly informative and can now collect spectral information from the crop canopy over a wide range of electromagnetic bands, which may then be translated into physiological and biochemical information about the crop canopy [
5]. Thus, field managers may now rely on remote-sensing technology to monitor crop growth.
Crop growth monitoring mainly traces crop growth status and variations in crop growth. Although crop growth is affected by many factors and the growth process is quite complex, it can be estimated using biochemical parameters such as biomass, leaf area index, and chlorophyll content. The crop canopy spectrum obtained by remote sensing technology provides further access to crop canopy biochemical information [
6]. The crop canopy spectrum is determined by the leaves, canopy structure, and soil background [
7]. Therefore, the relationship between spectral information and crop parameters can be established for estimating crop parameters, such as leaf area index (LAI), above-ground biomass, density, chlorophyll, plant nitrogen content, and photosynthetic pigments [
8,
9,
10,
11,
12].
Acquiring images from unmanned aerial vehicles (UAV) constitute a remote sensing technology that offers high resolution, high efficiency, rapidity, and low cost, which lead to more timely and accurate crop monitoring [
13,
14]. Its spatial resolution exceeds that of satellite-based remote sensing, and unlike ground remote sensing, it can generate orthophotos [
15,
16,
17]. Compared with traditional remote sensing, the flight pattern, time, maneuverability, and the cost of UAV remote sensing is more advantageous [
18]. UAV remote sensing technologies are increasingly used in agriculture to monitor crop growth and have achieved good results.
Vegetation indices, which are mathematical constructions involving the reflectance of different spectral bands, lead to more accurate information about vegetation [
8]. With the widespread application of remote sensing technology in agriculture, vegetation indices are often used to estimate crop parameters. Chen et al. [
19] used vegetation indices and a neural network algorithm to improve the estimation accuracy of maize leaf area index. Han et al. [
20] used four machine learning algorithms (multiple linear regression, support vector machine, artificial neural network, and random forest) to invert maize above-ground biomass (AGB) to improve the inversion effect. Swain et al. [
21] obtained high-resolution images of rice using an unmanned-helicopter, low-altitude, remote sensing platform to demonstrate that such images work well for estimating rice biomass. Chang et al. [
22] constructed a model to estimate corn chlorophyll content using the spectral vegetation indices and difference vegetation index. Schirrmann et al. [
23] used low-cost UAV images to monitor the physiological parameters and nitrogen content of wheat. Li et al. [
24] used four methods: partial least squares regression (PLSR), support vector machines (SVM), stepwise multiple linear regression (SMLR), and back-propagation neural network (BPN) to estimate the nitrogen content of winter wheat and used partial least squares and support vector machine to improve estimates. These studies all focused on empirical models, whereas others have studied semi-empirical models and physical models. For example, Duan et al. [
25] used the PROSAIL model to estimate the LAI of maize, potatoes, and sunflowers and showed that incorporating the direction information improves the estimation accuracy. In addition, Li et al. [
26] used the PROSPECT + SAIL model to invert the LAI of multiple crops with high inversion accuracy.
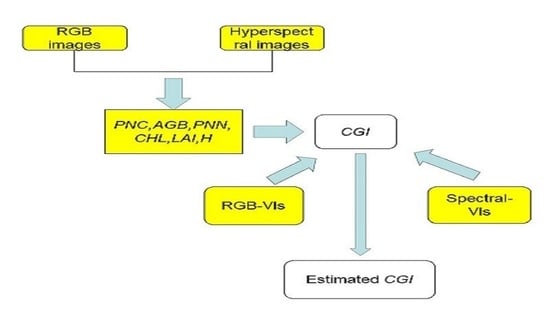
Crop growth is closely related to plant nitrogen content, above-ground biomass, plant water content, chlorophyll, LAI, plant height, and other factors [
15]. Therefore, to monitor crop growth, many studies use remote-sensing data to estimate a single parameter (e.g., LAI, AGB) related to crop growth and thereby determine growth status [
15]. The estimation of crop growth by a single growth parameter has been extensively studied; however, studies that combine multiple growth parameters to monitor crop growth have not been found. To explore the use of multiple crop-growth indicators to monitor crop growth, the present study uses UAV remote sensing data to monitor surface-scale crop growth. Specifically, we combine plant nitrogen content (
PNC), above-ground biomass (
AGB), plant water content (
PWC), chlorophyll (
CHL),
LAI, and plant height (
H) into a comprehensive growth index (
CGI). We take into account the different winter wheat growing stages when monitoring single growth phases versus the entire growth phase. More precisely, we evaluate crop-growth monitoring from the vegetation index based on UAV digital images and compare the results with those obtained from the vegetation-index-based UAV hyperspectral multi-temporal images. In addition, we combine multiple linear regression (MLR), partial least squares (PLSR), and random forest (RF) with the vegetation index to estimate the
CGI and map its spatial distribution.
The structure of this paper is as follows:
Section 2 presents the study area, the experimental design, the techniques used for ground sampling, data acquisition, and the processing of digital and hyperspectral remote-sensing data from UAVs. In addition, analytical methods, statistical methods, and vegetation indices are also discussed.
Section 3 discusses the selection of indices and how they affect the accuracy of the resulting
CGI. Specifically, we use the vegetation index based on UAV RGB imagery, the vegetation index based on UAV hyperspectral imagery, and the
PNC, AGB, PWC, CHL, LAI, and
H. We also contrast the use of only a single vegetation index with the use of multiple vegetation indices combined with MLR, PLSR, or RF.
Section 4 analyzes the advantages and disadvantages of the various methods and of the resulting estimates based on UAV RGB and hyperspectral remote sensing. Finally,
Section 5 discusses the potential applications of UAV RGB imagery and hyperspectral imagery in remote monitoring of agriculture.
2. Materials and Methods
2.1. Survey and Test Design of the Research Area
This study was conducted at the National Precision Agriculture Research and Demonstration Base of Xiaotangshan Town, Changping District, Beijing, China. It is located around in the area of the Yanshan branch vein and plain. The north latitude of this region is 40°00′–40°21′, and the east longitude is 116°34′–117°00′. The annual precipitation is approximately 645 mm. The highest and lowest temperatures can reach +40 and −10 °C, and the average temperature is 11.7 °C (from China Meteorological Data Service). The experimental field held a total of 48 plots. There are two winter wheat varieties, J9843 and ZM175, and three water treatments: 200, 100 mm, and rainfall. Water irrigation was applied on 20 October 2014, 9 April 2015, and 30 April 2015. To accentuate the differences in crop nitrogen content between the experimental plots, different amounts of nitrogen were supplied: each plot was provided with either 0(N1), 195(N2), 390(N3), or 585(N4) kg urea/hm
2. Each treatment scheme was repeated three times, and a total of 48 experimental plots were constructed.
Figure 1 shows the location of the test area and the experimental design.
2.2. Acquisition of Ground Data
We collected the PNC, AGB, PWC, CHL, LAI, and H data of winter wheat at (GS 31) (21 April 2015), (GS 47) (26 April 2015), and (GS 65) (13 May 2015). For the PNC, we used Buchi B-339 (Switzerland) and measured the nitrogen content of each organ (leaves, stems, and spikes) of 20 samples. We selected 20 plants in each growth period that represent the overall growth of the plot and investigated the wheat density of the plot to calculate the biomass of the plot. The dryness and freshness of the spikes in the flowering and filling stages of winter wheat were also considered. For AGB acquisition, 20 samples were taken from each growing area (6 × 8 m2), and stems and leaves were separated. The above-ground parts were heated to 105 °C in an oven for 30 min, then dried at 70 °C for about 24 h (i.e., until achieving a constant weight). The result is the dry mass per sampling area, which is the biomass. We calculated the PWC from the fresh and dry mass of sample stems, leaves, and ears. The total fresh quality was subtracted from the total dry quality which was then divided by the total fresh quality. Twenty leaves of different parts of the plant were randomly selected, and the leaf CHL content was measured by using a Dualex 4 nitrogen balance. Each leaf was measured five times, and the average value of the 20 leaves was used as the chlorophyll content of the sampling plot. To measure the LAI, 20 plant stems and leaves were separated. The leaf area was measured using a CID-203 Laser Leaf Area Meter (CID Company, Bethesda, MD, USA) to obtain the leaf area of a single stem. Next, the number of stems per unit area was determined through field investigation and multiplied by the total number of stems per unit area to calculate the LAI. Before the winter wheat began heading, a ruler was used to measure H from the stem base to the flag leaf tip. After heading, the distance from the stem base to the topmost end was measured.
2.3. UAV RGB-Data Acquisition and Processing
The experimental UAV remote sensing platform consisted of a DJI S1000 UAV (SZ DJI Technology Co., Ltd., Sham Chun, China) with eight rotors and equipped with two 18,000 mA h (25 V) batteries. It has 30 min of autonomy, its payload capacity is 6 kg, and its flight speed is 8 m/s. It was equipped with an RGB camera (Sony DSC–QX100, Sony, Tokyo, Japan) that weighed 0.179 kg and provided images of 5472 × 3648 pixels. The RGB images obtained were acquired under stable lighting conditions, so the flight time started after 12 a.m. (21 April 2015, 26 April 2015, and 13 May 2015), and the flight height was 80 m. The weather was clear with no wind and few clouds. High-resolution digital images were obtained of winter wheat at (GS 31), (GS 47), (GS 65), with a spatial resolution of 0.013 m. After obtaining the RGB imagery, we used Agisoft PhotoScan software (Agisoft PhotoScan Professional Pro, Version 1.1.6, Agisoft LLC, 11 Degtyarniy per., St. Petersburg, Russia, hereinafter referred to as PhotoScan) to stitch the RGB images. RGB image stitching requires input POS (position point altitude system) data. The POS data contained longitude, latitude, altitude, yaw angle, pitch angle, and rotation angle at the moment of image acquisition. To stitch the UAV RGB imagery, we used the POS data and RGB imagery from the UAV to restore the spatial attitude at the time of image capture and then generated sparse point clouds. A spatial grid was established based on the sparse point cloud, and ground control point information was added to optimize the spatial pose of the image to obtain a sparse point cloud with spatial information. We built a dense point cloud based on the sparse point cloud with spatial information to generate a three-dimensional polygon grid and construct spatial texture. This procedure allowed us to produce a high-definition digital orthophoto mosaic of the UAV flying area.
2.4. UAV Hyperspectral Data Acquisition and Processing
Hyperspectral data acquisition was carried using the FIREFLEYE imaging spectrometer (also known as UHD185, Germany). The UHD185 weighs 0.47 kg and covers a wavelength range from 450 to 950 nm. The hyperspectral data were sampled at 4 nm intervals, producing a 1000 × 1000 pixel image with 125 bands. Before the UAV hyperspectral flight, the UHD185 was calibrated using a black-and-white board. The flight height was 80 m. The acquired UAV hyperspectral images have a spatial resolution of 0.021 m, including the gray pixel with a spatial resolution of 0.01 m. The same flight routes were used to acquire remote-sensing images for the three winter wheat growth stages.
UAV hyperspectral data processing mainly involved image correction, image stitching, and extraction of reflectance. Image correction of hyperspectral images converts the digital number (DN) to the ground surface reflectance [
27].
The hyperspectral images have rich spectral information but lack texture information. For the stitching, we used the Cubert Cube-Pilot software (Cube-Pilot, Version 1.4, Cubert GmbH, Ulm, Baden-Württemberg, Germany) to fuse the hyperspectral images with the corresponding full-color image acquired at the same time to generate the fused hyperspectral image. We then used Agisoft PhotoScan software with the point cloud data from the full-color image to complete the stitching. The final spatial resolution of the hyperspectral image was about 5 cm [
28].
2.5. Research Methods
Specifically (see
Figure 2), we calculated the vegetation indices from both the UAV RGB imagery and from the UAV hyperspectral images and analyzed them both by comparison with
PNC, AGB, PWC, CHL, LAI, H, and
CGI. The
CGI estimation model was constructed using MLR, PLSR, and RF, and the maps of the CGI distribution based on the UAV-based RGB and spectral vegetation indices were generated.
2.6. Analysis Methods
We analyzed the correlation between the vegetation index and
CGI and used multiple MLR, PLSR, and RF methods to build an estimation model. To estimate the
CGI from a single vegetation index, to deal with multiple variables, we used MLR, PLSR, and RF. MLR can be used to accurately measure the degree of correlation between various factors and the degree of regression fitting to improve the prediction equation. The larger the absolute value of the standardized regression coefficient, the greater the effect of the corresponding independent variable on the dependent variable.
In Equation (1), n is the number of modeling factors and a (= 1, …, m) is the coefficient.
PLSR can effectively eliminate collinearity among multiple variables, reduce multiple variables to fewer unrelated latent variables, maximize covariance between independent and dependent variables, and then establish regression models [
29,
30]. RF is based on the bootstrap sampling method whereby multiple samples are extracted from the original sample. The number of random forest algorithm trees we use is set to 1000, and the number of nodes is 50. Each bootstrap sample is modeled using a decision tree, and multiple decision trees are then combined for prediction. Finally, the prediction is determined by voting [
31,
32].
We used WiMATLAB2018a software (Matrix Laboratory 2014a, MathWorks, Inc., Natick, MA, USA) to calibrate and verify the model with the vegetation index as the input variable and the CGI as the output variable.
2.7. Selection of RGB Imagery Indices and Hyperspectral Indices
The vegetation index combines two or more pieces of spectral information, which can simply the measurement of vegetation states. The vegetation index is widely used for monitoring grassland, forest, and drought. To build a model to estimate the
CGI, 13 RGB-imagery-based vegetation indices and 13 hyperspectral-image-based vegetation indices were selected (
Table 1).
2.8. Construction of Comprehensive Growth Index
In this study, we combine
PNC,
AGB,
PWC,
CHL,
LAI, and
H into a
CGI. We take into account the different winter wheat growth stages when monitoring single growth phases versus the entire growth phase. In order to combine multiple agronomic parameters into a new index and provide guidance for future remote sensing yield monitoring, the
PNC, AGB, PWC, CHL, LAI, and
H are combined into
CGI, which can not only reflect the growth information of crops but can also be correlated with yield. It is also of great significance to the monitoring of crops. Each parameter that constructs the CGI contributes to the construction of the model, so it is calculated in a weighted manner. For the time being, the contribution of each factor is the same; that is, the contribution of each factor to the construction of the model is the same. The
PNC, AGB, PWC, CHL, LAI, and
H are normalized separately:
After normalizing
PNC, AGB, PWC, CHL, LAI, and
H, they are weighted by one-sixth and summed to form the CGI [
15]:
where
t =
PNC, AGB, PWC, CHL, LAI, and
H;
is the value of
PNC, AGB, PWC, CHL, LAI, and
H at each the growth stage;
is the maximum of
PNC, AGB, PWC, CHL, LAI, and
H at each the growth stage;
is the normalized value; and
a, b, c, d, e, f are each one-sixth.
2.9. Verification of Accuracy
We took the 48 data from each of the three winter wheat growth stages, used two sets of data as calibration sets (plant field 1, plant field 2; see
Figure 1), and used the remaining set (plant field 3,
Figure 1) as the validation set for constructing the model. The correlation coefficient was used to evaluate how the vegetation indices were related to the
CGI. To evaluate the performance of the proposed model, we use the coefficient of determination
R2, the root-mean-squared error (
RMSE), and the normalized root-mean-squared error (
NRMSE). As a model evaluation standard [
15], statistically speaking, higher
R2 values and lower
RMSE and
NRMSE correspond to a more accurate model.
R2,
RMSE, and
NRMSE are calculated as follows:
where
is the measured
CGI for winter wheat,
is the average measured
CGI,
is the predicted
CGI,
is the average predicted
CGI, and
n is the number of model samples.
3. Results and Analysis
3.1. Correlation Analysis
To construct the proposed
CGI and vegetation index, the RGB imagery indices and spectral indices of (GS 31), (GS 47), (GS 65) and for all three stages were combined with
PNC, AGB, PWC, CHL, LAI, and
H.
Table 2 and
Table 3 list the correlation between
CGI, RGB imagery indices, and spectral indices. These results show that the correlation between the RGB imagery indices and
PNC, AGB, PWC, CHL, LAI, and
H depends on the growth period, and most of these correlations are very significant (significance level of 0.01).
We find that the RGB imagery indices r, EXR, VARI, and MGRVI correlate with the CGI during different growth stages, and the correlation coefficients are all higher than those between (i) r, EXR, VARI, MGRVI and (ii) PNC, AGB, PWC, CHL, LAI, and H. Thus, the correlation coefficients between RGB imagery indices r, EXR, VARI, MGRVI, and the CGI for the four growth stages are greater than the correlation coefficients between the individual crop-growth indicators. For the spectral indices, the correlation between the hyperspectral CGI and PNC, AGB, PWC, CHL, LAI, and H also hovers around 0.01. From (GS 31) to the total for the three stages, the correlation between the spectral index and the six crop-growth indicators varies irregularly. However, the spectral indices LCI, MSR, SR, and NDVI are more strongly correlated with the CGI than with the six crop-growth indicators. Thus, the CGI provides more accurate estimates of the crop-growth parameters.
3.2. Estimate of CGI Based on RGB Imagery and Spectral Indices
According to the results given in
Table 2 and
Table 3, we select the RGB-imagery indices r, EXR, VARI, and MGRVI and the spectral indices LCI, MSR, SR, and NDVI to estimate CGI.
The RGB-imagery indices r, EXR, VARI, and MGRVI and the spectral indices LCI, MSR, SR, and NDVI are taken as factors in (GS 31), (GS 47), (GS 65) and for all three stages, respectively. We then construct the CGI models based on the RGB-imagery indices r, EXR, VARI, and MGRVI and on the spectral indices LCI, MSR, SR, and NDVI for the different growth stages (see
Table 4).
The CGI model based on the RGB imagery indices r, EXR, VARI, and MGRVI generally performs well for all three growth stages and for the total of the three stages. The best performance for the RGB imagery index model is for the (GS 31). Overall, R2 is higher and the RMSE and NMRSE are lower. At the same time, several pairs of RGB imagery–index calibration sets have the same R2 and RMSE, such as VARI and MGRVI. The effect of the model becomes apparent only upon comparing NRMSE. The smaller the NRMSE, the higher the prediction accuracy. Of course, we also need to consider R2, RMSE, and NRMSE for the validation set. Similarly, the effects of EXR, VARI, and the two RGB imagery indices used to construct the CGI model during (GS 47) need to be analyzed by NRMSE. The CGI effect of each growth stage is compared. The best-performing RGB indices are r, r, r, and EXR.
In the estimation model based on the spectral indices LCI, MSR, SR, and NDVI, evaluating models based on different spectral indices also requires comparing the magnitudes of R2, RMSE, and NRMSE. We find that the difference between the R2 of different estimation models is relatively clear. Combining these spectral indices with the results of the validation model allows us to evaluate the performance of different spectral index models. A comparison of the results of each model shows that the estimation model based on LCI gives the most accurate results.
3.3. Using RGB VIs and Spectral VIs with Machine Learning to Estimate CGI
Table 5 shows the modeling analysis based on the four RGB imagery indices, the four spectral indices, and the
CGI estimated by using MLR, PLSR, and RF.
Comparing the estimation of the
CGI result based on indices built from RGB imagery with that based on the spectral indices from the different growth stages, we find that the latter is superior to the former. In addition, a comparison with different machine learning methods shows that the
CGI estimation model constructed using MLR is the most accurate of the three methods, followed by the model constructed from PLSR, and finally by the model constructed from RF, which shows that MLR method is the index, followed by PLSR, and finally by RF. We now use the verification set data to verify the RGB-imagery -based index and spectral-based index combined with the MLR, PLSR, and RF methods to estimate the
CGI.
Figure 3 and
Figure 4 show the relationship between the measured and predicted values (y = ax + b,
R2,
RMSE,
NRMSE). The results show that the verification is consistent with the modeling, and the fit is very good (the fit to the spectral index is better than the fit to the RGB-imagery index). Comprehensive modeling and verification (see
Table 5 and
Figure 3 and
Figure 4) show that the MLR, PLSR, and RF methods all provide a more accurate estimate of the
CGI than the single RGB-imagery index or the single spectral index (see
Table 4 and
Table 5 and
Figure 3 and
Figure 4). Thus, these three methods all provide good estimates of the
CGI.
3.4. Map of CGI Distribution
Estimating the
CGI by using machine learning and based on RGB-imagery-based indices and spectral-based indices allows us to select the best estimation model for the different growth stages.
Figure 5 and
Figure 6 show the results of applying the MLR estimation model to the UAV RGB and hyperspectral imagery of (GS 31), (GS 47), and (GS 65).
From jointing to flowering, the average RGB-based
CGI ranges from 0.74 to 0.79 (
Figure 5) and hyperspectral-based CGI averages from 0.74 to 0.78 (
Figure 6). In addition, the corresponding
CGI map is greener, indicating a higher
CGI. The current map also shows that the
CGI has increased during the three growing seasons of winter wheat. These results are consistent with the actual observations. The
CGI distribution based on the RGB and hyperspectral UAV images shows the difference between
CGI estimation models based on the two types of data (
Figure 7). The predicted
CGI values differ, which leads to different
CGI distributions for the same growth stage. However, the results for the three growth stages show that winter wheat growth is best in plant field 2 and is relatively stable over the different growth stages. The map of the
CGI distribution clearly differentiates between the better-growing areas and the poorer-growing areas.
5. Conclusions
Six growth indicators were used to construct a new CGI index. The CGI of winter wheat at different growth stages was estimated by using UAV RGB imagery and hyperspectral data. The linear, nonlinear, MLR, PLSR, and RF models were further developed. The main conclusions are as follows: The RGB imagery indices r, EXR, VARI, and MGRVI are more strongly correlated with the CGI for the growth stages than are PNC, AGB, PWC, CHL, LAI, and H. The spectral indices LCI, MSR, SR, and NDVI are more strongly correlated with the CGI than the single growth-monitoring index for the four stages, which indicates that the CGI can be used to monitor crop growth. On the one hand, the CGI model from a single RGB imagery index has different optimal indices at different growth stages. On the other hand, the model constructed by using a single spectral index has the best index in each growth stage, all of which are in terms of the LCI. By combining multiple RGB imagery indices and spectral indices with MLR, PLSR, and RF to estimate the CGI, the MLR model is optimal for different growth stages, the PLSR model is second best, and the RF model is the worst. The best model based on RGB-imagery indices gives R2, RMSE, and NRMSE of 0.73, 0.04, 5.69%, and the best model based on hyperspectral data gives an R2, RMSE, and NRMSE of 0.78, 0.04, and 5.29%, respectively. The CGI can be accurately estimated by using UAV RGB imagery and hyperspectral data. Both methods can estimate the CGI. However, the hyperspectral data lead to more accurate estimates of the CGI than the RGB imagery, which demonstrates that UAV hyperspectral imaging provides more accurate results. Currently, developments are underway for UAV payloads, including hyperspectral, LIDAR, synthetic aperture radars, and thermal infrared sensors. Fully comparing the advantages of each sensor type requires further study.