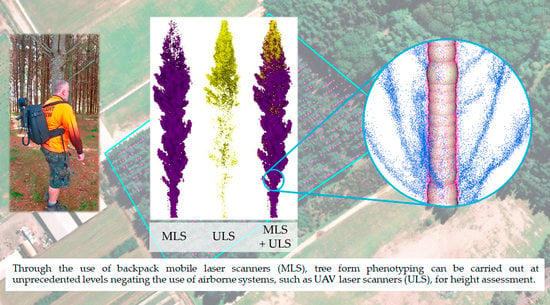
Assessing the Potential of Backpack-Mounted Mobile Laser Scanning Systems for Tree Phenotyping
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Capture
2.1.1. Study Site
2.1.2. Field Data
2.1.3. MLS Data
2.1.4. ULS Data
2.1.5. Ground Control: CloudReg
2.2. Data Processing and Analysis
2.2.1. MLS Data Processing
2.2.2. ULS Data Processing
2.2.3. Individual Tree Segmentation
2.2.4. Derived Tree Form Metrics
2.2.5. Accuracy Statistics
3. Results
3.1. Individual Tree Segmentation
3.2. Tree Metrics
3.2.1. DBH
3.2.2. Tree Height
3.2.3. Stem Volume
3.2.4. Whorl Detection
4. Discussion
4.1. Individual Tree Segmentation
4.2. Tree Metrics
4.2.1. DBH
4.2.2. Height
4.2.3. Stem Volume
4.2.4. Whorl Detection
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Costa, C.; Schurr, U.; Loreto, F.; Menesatti, P.; Carpentier, S. Plant phenotyping research trends, a science mapping approach. Front. Plant Sci. 2018, 9, 1933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dungey, H.S.; Dash, J.P.; Pont, D.; Clinton, P.W.; Watt, M.S.; Telfer, E.J. Phenotyping whole forests will help to track genetic performance. Trends Plant Sci. 2018, 23, 854–864. [Google Scholar] [CrossRef] [PubMed]
- Pont, D.; Dungey, H.S.; Suontama, M.; Stovold, G.T. Spatial Models with Inter-Tree Competition from Airborne Laser Scanning Improve Estimates of Genetic Variance. Front. Plant Sci. 2020, 11, 596315. [Google Scholar] [CrossRef] [PubMed]
- Bombrun, M.; Dash, J.P.; Pont, D.; Watt, M.S.; Pearse, G.D.; Dungey, H.S. Forest-scale phenotyping: Productivity characterisation through machine learning. Front. Plant Sci. 2020, 11, 99. [Google Scholar] [CrossRef]
- Hartley, R.J.; Leonardo, E.M.; Massam, P.; Watt, M.S.; Estarija, H.J.; Wright, L.; Melia, N.; Pearse, G.D. An assessment of high-density UAV point clouds for the measurement of young forestry trials. Remote Sens. 2020, 12, 4039. [Google Scholar] [CrossRef]
- Pearse, G.D.; Watt, M.S.; Dash, J.P.; Stone, C.; Caccamo, G. Comparison of models describing forest inventory attributes using standard and voxel-based lidar predictors across a range of pulse densities. Int. J. Appl. Earth Obs. Geoinf. 2019, 78, 341–351. [Google Scholar] [CrossRef]
- Pont, D. Assessment of Individual Trees Using Aerial Laser Scanning in New Zealand Radiata Pine Forests. Ph.D. Thesis, University of Canterbury, Christchurch, New Zealand, 2016. Available online: https://hdl.handle.net/10092/12872 (accessed on 5 November 2021).
- Watt, M.S.; Meredith, A.; Watt, P.; Gunn, A. The influence of LiDAR pulse density on the precision of inventory metrics in young unthinned Douglas-fir stands during initial and subsequent LiDAR acquisitions. N. Z. J. For. Sci. 2014, 44, 18. [Google Scholar] [CrossRef] [Green Version]
- Windrim, L.; Bryson, M. Detection, Segmentation, and Model Fitting of Individual Tree Stems from Airborne Laser Scanning of Forests Using Deep Learning. Remote Sens. 2020, 12, 1469. [Google Scholar] [CrossRef]
- Kellner, J.R.; Armston, J.; Birrer, M.; Cushman, K.; Duncanson, L.; Eck, C.; Falleger, C.; Imbach, B.; Král, K.; Krůček, M. New opportunities for forest remote sensing through ultra-high-density drone lidar. Surv. Geophys. 2019, 40, 959–977. [Google Scholar] [CrossRef] [Green Version]
- Thies, M.; Pfeifer, N.; Winterhalder, D.; Gorte, B.G. Three-dimensional reconstruction of stems for assessment of taper, sweep and lean based on laser scanning of standing trees. Scand. J. For. Res. 2004, 19, 571–581. [Google Scholar] [CrossRef]
- Hopkinson, C.; Chasmer, L.; Young-Pow, C.; Treitz, P. Assessing forest metrics with a ground-based scanning lidar. Can. J. For. Res. 2004, 34, 573–583. [Google Scholar] [CrossRef] [Green Version]
- Lovell, J.; Jupp, D.L.; Culvenor, D.; Coops, N. Using airborne and ground-based ranging lidar to measure canopy structure in Australian forests. Can. J. Remote Sens. 2003, 29, 607–622. [Google Scholar] [CrossRef]
- Watt, P.J.; Donoghue, D.N.M. Measuring forest structure with terrestrial laser scanning. Int. J. Remote Sens. 2005, 26, 1437–1446. [Google Scholar] [CrossRef]
- Kankare, V.; Joensuu, M.; Vauhkonen, J.; Holopainen, M.; Tanhuanpää, T.; Vastaranta, M.; Hyyppä, J.; Hyyppä, H.; Alho, P.; Rikala, J.; et al. Estimation of the timber quality of scots pine with terrestrial laser scanning. Forests 2014, 5, 1879–1895. [Google Scholar] [CrossRef] [Green Version]
- Abegg, M.; Kükenbrink, D.; Zell, J.; Schaepman, M.E.; Morsdorf, F. Terrestrial laser scanning for forest inventories-tree diameter distribution and scanner location impact on occlusion. Forests 2017, 8, 184. [Google Scholar] [CrossRef] [Green Version]
- Mengesha, T.; Hawkins, M.; Tarleton, M.; Nieuwenhuis, M. Stem quality assessment using terrestrial laser scanning technology: A case study of ash trees with a range of defects in two stands in Ireland. Scand. J. For. Res. 2015, 30, 605–616. [Google Scholar] [CrossRef]
- Liang, X.; Kankare, V.; Hyyppä, J.; Wang, Y.; Kukko, A.; Haggrén, H.; Yu, X.; Kaartinen, H.; Jaakkola, A.; Guan, F.; et al. Terrestrial laser scanning in forest inventories. ISPRS J. Photogramm. Remote Sens. 2016, 115, 63–77. [Google Scholar] [CrossRef]
- Newnham, G.J.; Armston, J.D.; Calders, K.; Disney, M.I.; Lovell, J.L.; Schaaf, C.B.; Strahler, A.H.; Danson, F.M. Terrestrial laser scanning for plot-scale forest measurement. Curr. For. Rep. 2015, 1, 239–251. [Google Scholar] [CrossRef] [Green Version]
- Raumonen, P.; Åkerblom, M.; Kaasalainen, M.; Casella, E.; Calders, K.; Murphy, S. Massive-scale tree modelling from TLS data. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2015, 2, 189–196. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Hyyppä, J.; Kaartinen, H.; Lehtomäki, M.; Pyörälä, J.; Pfeifer, N.; Holopainen, M.; Brolly, G.; Francesco, P.; Hackenberg, J.; et al. International benchmarking of terrestrial laser scanning approaches for forest inventories. ISPRS J. Photogramm. Remote Sens. 2018, 144, 137–179. [Google Scholar] [CrossRef]
- Cabo, C.; Del Pozo, S.; Rodríguez-Gonzálvez, P.; Ordóñez, C.; González-Aguilera, D. Comparing terrestrial laser scanning (TLS) and wearable laser scanning (WLS) for individual tree modeling at plot level. Remote Sens. 2018, 10, 540. [Google Scholar] [CrossRef] [Green Version]
- Holopainen, M.; Kankare, V.; Vastaranta, M.; Liang, X.; Lin, Y.; Vaaja, M.; Yu, X.; Hyyppä, J.; Hyyppä, H.; Kaartinen, H. Tree mapping using airborne, terrestrial and mobile laser scanning–A case study in a heterogeneous urban forest. Urban For. Urban Green. 2013, 12, 546–553. [Google Scholar] [CrossRef]
- Bauwens, S.; Bartholomeus, H.; Calders, K.; Lejeune, P. Forest inventory with terrestrial LiDAR: A comparison of static and hand-held mobile laser scanning. Forests 2016, 7, 127. [Google Scholar] [CrossRef] [Green Version]
- Ryding, J.; Williams, E.; Smith, M.J.; Eichhorn, M.P. Assessing handheld mobile laser scanners for forest surveys. Remote Sens. 2015, 7, 1095–1111. [Google Scholar] [CrossRef] [Green Version]
- Hyyppä, E.; Yu, X.; Kaartinen, H.; Hakala, T.; Kukko, A.; Vastaranta, M.; Hyyppä, J. Comparison of backpack, handheld, under-canopy UAV, and above-canopy UAV laser scanning for field reference data collection in boreal forests. Remote Sens. 2020, 12, 3327. [Google Scholar] [CrossRef]
- Shao, J.; Zhang, W.; Mellado, N.; Wang, N.; Jin, S.; Cai, S.; Luo, L.; Lejemble, T.; Yan, G. SLAM-aided forest plot mapping combining terrestrial and mobile laser scanning. ISPRS J. Photogramm. Remote Sens. 2020, 163, 214–230. [Google Scholar] [CrossRef]
- Bienert, A.; Georgi, L.; Kunz, M.; Maas, H.G.; von Oheimb, G. Comparison and combination of mobile and terrestrial laser scanning for natural forest inventories. Forests 2018, 8, 395. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.; Chen, Y.; Kukko, A.; Kaartinen, H.; Jaakkola, A.; Khoramshahi, E.; Hakala, T.; Hyyppä, J.; Holopainen, M.; Hyyppä, H. SLAM-aided stem mapping for forest inventory with small-footprint mobile LiDAR. Forests 2015, 6, 4588–4606. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Wan, P.; Wang, T.; Cai, S.; Chen, Y.; Jin, X.; Yan, G. A novel approach for the detection of standing tree stems from plot-level terrestrial laser scanning data. Remote Sens. 2019, 11, 211. [Google Scholar] [CrossRef] [Green Version]
- Bu, G.; Wang, P. Adaptive circle-ellipse fitting method for estimating tree diameter based on single terrestrial laser scanning. J. Appl. Remote Sens. 2016, 10, 026040. [Google Scholar] [CrossRef]
- Buck, A.L.B.; Lingnau, C.; Neto, S.P.; Machado, Á.M.L.; Martins-Neto, R.P. Stem modelling of eucalyptus by terrestrial laser scanning. Floresta E Ambiente 2019, 26, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Boudon, F.; Preuksakarn, C.; Ferraro, P.; Diener, J.; Nacry, P.; Nikinmaa, E.; Godin, C. Quantitative assessment of automatic reconstructions of branching systems obtained from laser scanning. Ann. Bot. 2014, 114, 853–862. [Google Scholar] [CrossRef] [Green Version]
- Bruggisser, M.; Hollaus, M.; Otepka, J.; Pfeifer, N. Influence of ULS acquisition characteristics on tree stem parameter estimation. ISPRS J. Photogramm. Remote Sens. 2020, 168, 28–40. [Google Scholar] [CrossRef]
- Torresan, C.; Carotenuto, F.; Chiavetta, U.; Miglietta, F.; Zaldei, A.; Gioli, B. Individual tree crown segmentation in two-layered dense mixed forests from uav lidar data. Drones 2020, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Dash, J.P.; Watt, M.S.; Hartley, R.J.L. Testing UAV-Borne Riegl Mini VUX-1 Scanner for Phenotyping a Mature Genetics Trial. In Technical Notes from the Growing Confidence in Forestry’s Future Research Programme; Scion: Rotorua, New Zealand, 2019; Volume TN-023, Available online: https://gcff.nz/publications/technical-notes/ (accessed on 1 May 2020).
- Escandón, M.; Valledor, L.; Pascual, J.; Pinto, G.; Cañal, M.J.; Meijón, M. System-wide analysis of short-term response to high temperature in Pinus radiata. J. Exp. Bot. 2017, 68, 3629–3641. [Google Scholar] [CrossRef]
- Yan, H.; Bi, H.; Li, R.; Eldridge, R.; Wu, Z.; Li, Y.; Simpson, J. Assessing climatic suitability of Pinus radiata (D. Don) for summer rainfall environment of southwest China. For. Ecol. Manag. 2006, 234, 199–208. [Google Scholar] [CrossRef]
- Cabo, C.; Ordóñez, C.; López-Sánchez, C.A.; Armesto, J. Automatic dendrometry: Tree detection, tree height and diameter estimation using terrestrial laser scanning. Int. J. Appl. Earth Obs. Geoinf. 2018, 69, 164–174. [Google Scholar] [CrossRef]
- Rais, A.; Poschenrieder, W.; Pretzsch, H.; van de Kuilen, J.-W.G. Influence of initial plant density on sawn timber properties for Douglas-fir (Pseudotsuga menziesii (Mirb.) Franco). Ann. For. Sci. 2014, 71, 617–626. [Google Scholar] [CrossRef]
- Grace, J.; Pont, D.; Goulding, C.; Rawley, B. Modelling branch development for forest management. N. Z. J. For. Sci. 1999, 29, 391–408. [Google Scholar]
- Dassot, M.; Colin, A.; Santenoise, P.; Fournier, M.; Constant, T. Terrestrial laser scanning for measuring the solid wood volume, including branches, of adult standing trees in the forest environment. Comput. Electron. Agric. 2012, 89, 86–93. [Google Scholar] [CrossRef]
- Du, S.; Lindenbergh, R.; Ledoux, H.; Stoter, J.; Nan, L. AdTree: Accurate, detailed, and automatic modelling of laser-scanned trees. Remote Sens. 2019, 11, 2074. [Google Scholar] [CrossRef] [Green Version]
- Fang, R.; Strimbu, B.M. Comparison of mature Douglas-firs’ crown structures developed with two quantitative structural models using TLS point clouds for neighboring trees in a natural regime stand. Remote Sens. 2019, 11, 1661. [Google Scholar] [CrossRef] [Green Version]
- Pyörälä, J.; Liang, X.; Vastaranta, M.; Saarinen, N.; Kankare, V.; Wang, Y.; Holopainen, M.; Hyyppä, J. Quantitative assessment of Scots pine (Pinus sylvestris L.) whorl structure in a forest environment using terrestrial laser scanning. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 3598–3607. [Google Scholar] [CrossRef] [Green Version]
- Harikumar, A.; Bovolo, F.; Xinlian, L. An effective approach to 3D stem modeling and branch-knot localization in multiscan TLS data. In Proceedings of the International Geoscience and Remote Sensing Symposium (IGARSS), Yokohama, Japan, 28 July–2 August 2019; pp. 6075–6078. [Google Scholar]
- Stängle, S.M.; Brüchert, F.; Kretschmer, U.; Spiecker, H.; Sauter, U.H. Clear wood content in standing trees predicted from branch scar measurements with terrestrial LiDAR and verified with X-ray computed tomography. Can. J. For. Res. 2014, 44, 145–153. [Google Scholar] [CrossRef]
- CNI Regional YTGEN User Group. PlotSafe: Overlapping Feature Cruising. Forest Inventory Procedures; CNI Regional YTGEN User Group: Rotorua, New Zealand, 2007. [Google Scholar]
- Wallace, L.O.; Lucieer, A.; Watson, C.S. Assessing the feasibility of UAV-based lidar for high resolution forest change detection. In Proceedings of the International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences-ISPRS Archives, Melbourne, Australia, 25 August–1 September 2012; pp. 499–504. [Google Scholar]
- Hackenberg, J.; Calders, K.; Demol, M.; Raumonen, P.; Piboule, A.; Disney, M. SimpleForest-a comprehensive tool for 3d reconstruction of trees from forest plot point clouds. BioRxiv 2021. [Google Scholar] [CrossRef]
- Dijkstra, E.W. A note on two problems in connexion with graphs. Numer. Math. 1959, 1, 269–271. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- De Conto, T.; Olofsson, K.; Görgens, E.B.; Rodriguez, L.C.E.; Almeida, G. Performance of stem denoising and stem modelling algorithms on single tree point clouds from terrestrial laser scanning. Comput. Electron. Agric. 2017, 143, 165–176. [Google Scholar] [CrossRef]
- Roussel, J.-R.; Auty, D.; De Boissieu, F.; Meador, A. lidR: Airborne LiDAR Data Manipulation and Visualization for Forestry Applications, R Package Version; R Foundation for Statistical Computing: Vienna, Austria, 2018; Volume 1. [Google Scholar]
- Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Zhong, L.; Cheng, L.; Xu, H.; Wu, Y.; Chen, Y.; Li, M. Segmentation of individual trees from TLS and MLS data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 774–787. [Google Scholar] [CrossRef]
- Bienert, A.; Georgi, L.; Kunz, M.; von Oheimb, G.; Maas, H.-G. Automatic extraction and measurement of individual trees from mobile laser scanning point clouds of forests. Ann. Bot. 2021, 128, 787–804. [Google Scholar] [CrossRef]
- Giannetti, F.; Puletti, N.; Quatrini, V.; Travaglini, D.; Bottalico, F.; Corona, P.; Chirici, G. Integrating terrestrial and airborne laser scanning for the assessment of single-tree attributes in Mediterranean forest stands. Eur. J. Remote Sens. 2018, 51, 795–807. [Google Scholar] [CrossRef] [Green Version]
- Calders, K.; Newnham, G.; Burt, A.; Murphy, S.; Raumonen, P.; Herold, M.; Culvenor, D.; Avitabile, V.; Disney, M.; Armston, J.; et al. Nondestructive estimates of above-ground biomass using terrestrial laser scanning. Methods Ecol. Evol. 2015, 6, 198–208. [Google Scholar] [CrossRef]
- Chen, S.; Feng, Z.; Chen, P.; Khan, T.U.; Lian, Y. Nondestructive estimation of the above-ground biomass of multiple tree species in boreal forests of china using terrestrial laser scanning. Forests 2019, 10, 936. [Google Scholar] [CrossRef] [Green Version]
- Čerňava, J.; Tuček, J.; Koreň, M.; Mokroš, M. Estimation of diameter at breast height from mobile laser scanning data collected under a heavy forest canopy. J. For. Sci. 2017, 63, 433–441. [Google Scholar] [CrossRef]
- Brolly, G.; Király, G. Algorithms for stem mapping by means of terrestrial laser scanning. Acta Silv. Et Lignaria Hung. 2009, 5, 119–130. [Google Scholar]
- Fleck, S.; Mölder, I.; Jacob, M.; Gebauer, T.; Jungkunst, H.F.; Leuschner, C. Comparison of conventional eight-point crown projections with LIDAR-based virtual crown projections in a temperate old-growth forest. Ann. For. Sci. 2011, 68, 1173–1185. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Li, Z.; Gong, P.; Cheng, X.; Clinton, N.; Cao, C.; Ni, W.; Wang, L. Automated methods for measuring DBH and tree heights with a commercial scanning lidar. Photogramm. Eng. Remote Sens. 2011, 77, 219–227. [Google Scholar] [CrossRef]
- Liang, X.; Hyyppä, J. Automatic stem mapping by merging several terrestrial laser scans at the feature and decision levels. Sensors 2013, 13, 1614–1634. [Google Scholar] [CrossRef] [Green Version]
- Maas, H.G.; Bienert, A.; Scheller, S.; Keane, E. Automatic forest inventory parameter determination from terrestrial laser scanner data. Int. J. Remote Sens. 2008, 29, 1579–1593. [Google Scholar] [CrossRef]
- Moskal, L.M.; Zheng, G. Retrieving forest inventory variables with terrestrial laser scanning (TLS) in urban heterogeneous forest. Remote Sens. 2012, 4, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Puliti, S.; Dash, J.P.; Watt, M.S.; Breidenbach, J.; Pearse, G.D. A comparison of UAV laser scanning, photogrammetry and airborne laser scanning for precision inventory of small-forest properties. Forestry 2020, 93, 150–162. [Google Scholar] [CrossRef]
- Sankey, T.; Donager, J.; McVay, J.; Sankey, J.B. UAV lidar and hyperspectral fusion for forest monitoring in the southwestern USA. Remote Sens. Environ. 2017, 195, 30–43. [Google Scholar] [CrossRef]
- Corte, A.P.D.; Rex, F.E.; de Almeida, D.R.A.; Sanquetta, C.R.; Silva, C.A.; Moura, M.M.; Wilkinson, B.; Zambrano, A.M.A.; da Cunha Neto, E.M.; Veras, H.F.P.; et al. Measuring individual tree diameter and height using gatoreye high-density UAV-lidar in an integrated crop-livestock-forest system. Remote Sens. 2020, 12, 863. [Google Scholar] [CrossRef] [Green Version]
- Jaakkola, A.; Hyyppä, J.; Yu, X.; Kukko, A.; Kaartinen, H.; Liang, X.; Hyyppä, H.; Wang, Y. Autonomous collection of forest field reference—The outlook and a first step with UAV laser scanning. Remote Sens. 2017, 9, 785. [Google Scholar] [CrossRef] [Green Version]
- Camarretta, N.; Harrison, P.A.; Lucieer, A.; Potts, B.M.; Davidson, N.; Hunt, M. From drones to phenotype: Using UAV-LiDAR to detect species and provenance variation in tree productivity and structure. Remote Sens. 2020, 12, 3184. [Google Scholar] [CrossRef]
- Liang, X.; Wang, Y.; Pyörälä, J.; Lehtomäki, M.; Yu, X.; Kaartinen, H.; Kukko, A.; Honkavaara, E.; Issaoui, A.E.I.; Nevalainen, O.; et al. Forest in situ observations using unmanned aerial vehicle as an alternative of terrestrial measurements. For. Ecosyst. 2019, 6, 20. [Google Scholar] [CrossRef] [Green Version]
- Ayrey, E.; Fraver, S.; Kershaw, J.A., Jr.; Kenefic, L.S.; Hayes, D.; Weiskittel, A.R.; Roth, B.E. Layer stacking: A novel algorithm for individual forest tree segmentation from LiDAR point clouds. Can. J. Remote Sens. 2017, 43, 16–27. [Google Scholar] [CrossRef]
- Wang, Y.; Lehtomäki, M.; Liang, X.; Pyörälä, J.; Kukko, A.; Jaakkola, A.; Liu, J.; Feng, Z.; Chen, R.; Hyyppä, J. Is field-measured tree height as reliable as believed–A comparison study of tree height estimates from field measurement, airborne laser scanning and terrestrial laser scanning in a boreal forest. ISPRS J. Photogramm. Remote Sens. 2019, 147, 132–145. [Google Scholar] [CrossRef]
- Larjavaara, M.; Muller-Landau, H.C. Measuring tree height: A quantitative comparison of two common field methods in a moist tropical forest. Methods Ecol. Evol. 2013, 4, 793–801. [Google Scholar] [CrossRef]
- Saarinen, N.; Kankare, V.; Vastaranta, M.; Luoma, V.; Pyörälä, J.; Tanhuanpää, T.; Liang, X.; Kaartinen, H.; Kukko, A.; Jaakkola, A. Feasibility of Terrestrial laser scanning for collecting stem volume information from single trees. ISPRS J. Photogramm. Remote Sens. 2017, 123, 140–158. [Google Scholar] [CrossRef]
- Murphy, G. Determining stand value and log product yields using terrestrial lidar and optimal bucking: A case study. J. For. 2008, 106, 317–324. [Google Scholar]
- Brede, B.; Calders, K.; Lau, A.; Raumonen, P.; Bartholomeus, H.M.; Herold, M.; Kooistra, L. Non-destructive tree volume estimation through quantitative structure modelling: Comparing UAV laser scanning with terrestrial LIDAR. Remote Sens. Environ. 2019, 233, 111355. [Google Scholar] [CrossRef]
- Wang, D.; Hollaus, M.; Puttonen, E.; Pfeifer, N. Automatic and self-adaptive stem reconstruction in landslide-affected forests. Remote Sens. 2016, 8, 974. [Google Scholar] [CrossRef] [Green Version]



















| No. Trees | DBH Range (cm) | Mean DBH (SD) (cm) | Height Range (m) | Mean Height (SD) (m) |
|---|---|---|---|---|
| 884 | 2.2–67.1 | 30.5 (12.58) | 1.7–34.4 | 23.9 (7.93) |
| Variables | R2 | RMSE (m) | RMSE (%) | MBE (m) |
|---|---|---|---|---|
| ULS vs. Field | 0.24 | 7.19 | 28.60 | −3.57 |
| ULS vs. Field (minus suppressed) | 0.42 | 2.85 | 10.14 | −1.32 |
| MLS vs. Field | 0.22 | 7.19 | 28.57 | −3.46 |
| MLS vs. Field (minus suppressed) | 0.41 | 2.78 | 9.90 | −1.18 |
| ULS vs. MLS | 0.94 | 0.87 | 3.02 | 0.11 |
| Individual Tree Identifier | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A8 | B23 | C7 | D14 | E8 | F19 | G18 | H7 | J5 | K1 | L8 | M7 | |
| Max Ht (m) | 13.9 | 14.0 | 10.5 | 10.10 | 17.9 | 10.8 | 12.9 | 9.1 | 17.1 | 15.8 | 18.9 | 17.9 |
| Stem volume (m3) | 2.77 | 1.59 | 0.99 | 0.97 | 2.15 | 1.03 | 1.27 | 0.69 | 2.69 | 3.80 | 3.90 | 2.80 |
| Max Ht Whorl (5 cm) | 15.38 | 16.03 | 13.18 | 16.38 | 17.63 | 16.68 | 16.73 | 14.73 | 13.78 | 15.43 | 18.48 | 17.48 |
| Max Ht Whorl (10 cm) | 16.1 | 16.9 | 15.4 | 18.1 | 17.5 | 18.8 | 18.1 | 13.51 | 17.1 | 15.9 | 18.6 | 17.6 |
| Max Ht Whorl (20 cm) | 16.15 | 17.15 | 15.15 | 19.15 | 18.75 | 18.95 | 18.75 | 15.16 | 17.15 | 16.35 | 18.55 | 19.35 |
| Tuning (cm) | No. Whorls Measured | No. Whorls Detected | No. True Positives | No. False Positives | No. False Negatives | RMSE (m) | RMSE (%) | MBE (m) | Detection Accuracy (%) |
|---|---|---|---|---|---|---|---|---|---|
| 5 | 410 | 602 | 289 | 313 | 108 | 0.17 | 1.88 | −0.01 | 40.25 |
| 10 | 410 | 265 | 201 | 64 | 209 | 0.26 | 2.73 | −0.01 | 42.41 |
| 20 | 410 | 193 | 175 | 18 | 235 | 0.22 | 2.10 | −0.01 | 40.89 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hartley, R.J.L.; Jayathunga, S.; Massam, P.D.; De Silva, D.; Estarija, H.J.; Davidson, S.J.; Wuraola, A.; Pearse, G.D. Assessing the Potential of Backpack-Mounted Mobile Laser Scanning Systems for Tree Phenotyping. Remote Sens. 2022, 14, 3344. https://doi.org/10.3390/rs14143344
Hartley RJL, Jayathunga S, Massam PD, De Silva D, Estarija HJ, Davidson SJ, Wuraola A, Pearse GD. Assessing the Potential of Backpack-Mounted Mobile Laser Scanning Systems for Tree Phenotyping. Remote Sensing. 2022; 14(14):3344. https://doi.org/10.3390/rs14143344
Chicago/Turabian StyleHartley, Robin J. L., Sadeepa Jayathunga, Peter D. Massam, Dilshan De Silva, Honey Jane Estarija, Sam J. Davidson, Adedamola Wuraola, and Grant D. Pearse. 2022. "Assessing the Potential of Backpack-Mounted Mobile Laser Scanning Systems for Tree Phenotyping" Remote Sensing 14, no. 14: 3344. https://doi.org/10.3390/rs14143344