1. Introduction
Smallholder farms with small plots (typically ≤2 ha) and complex cropping practices are the most common and important forms of agriculture worldwide, accounting for approximately 87% of the world’s existing agricultural land and producing 70–80% of the world’s food [
1,
2]. Although smallholder farming systems vary greatly in different countries and agricultural regions, they are generally characterized by limited farmland, decentralized management, and a low input-output ratio [
3,
4,
5,
6]. The existence of these characteristics makes these systems particularly vulnerable to global climate and environmental changes, explosive population growth, and market turmoil, posing serious threats to community food security and sustainable livelihoods [
7,
8,
9]. In this context, timely and accurate mapping and monitoring of crop patterns on smallholder farms are critical for scientists and planners in developing effective strategies to address these threats [
5,
6,
10]. Recent advances in high-resolution earth observations have provided new possibilities for mapping complex smallholder agricultural landscapes [
11,
12,
13].
Fine-scale remote sensing (RS) mapping of smallholder crops remains challenging, due to the high fragmentation and heterogeneity of agricultural landscapes. Over the past decade, many studies have been conducted using RS technology to objectively identify and map crop types and planting intensity at national, regional, and other spatial scales [
14,
15,
16,
17,
18]. Despite its diverse uses, RS technology has not been widely used in parcel-level smallholder crop mapping, which is critical to better predicting grain yields and determining area-based subsidies [
19,
20,
21]. Especially for some developing countries dominated by smallholder agriculture, such as Bangladesh and China, RS is urgently needed to establish their own parcel-level crop identification systems to support the development of local precision agriculture [
5,
22]. However, the following complex practical conditions render the application of RS technology in this particular field extremely challenging. First, parcels on smallholder farms are generally small and are accompanied by complex planting patterns. Second, multiple crops are planted in one area, and even the same crop can be planted and harvested on different dates. Third, there might be intercropping and mixed cropping, resulting in more than one crop being planted in the same parcel in the same season. The aforementioned complex factors make traditional medium- or low-spatial-resolution satellite images, such as those from the Moderate Resolution Imaging Spectroradiometer (MODIS) and Landsat sensors, unreliable for fine-scale mapping of crop planting patterns on smallholder farms [
16]. Fortunately, various very high spatial resolution (VHSR) satellite platforms (e.g., WorldView, Gaofen, and RapidEye) have emerged that can provide meter-level and even submeter-level resolutions, making it possible to map parcel-level smallholder crops [
23,
24].
VHSR images allow one to use geographic object-based image analysis (GEOBIA) technology [
25,
26], thus paving the way for parcel-level mapping of smallholder crops. In VHSR image analysis, the object to be identified is generally much larger than pixel size [
27,
28]. Unlike pixel-based methods, GEOBIA treats the basic target unit as an image object rather than a single pixel, more in line with the requirements of VHSR image analysis [
12,
26,
29]. Furthermore, GEOBIA performs image segmentation to construct a polygon network of homogeneous objects that, in the case of crop classification, matches the parcel boundaries [
27,
30]. Although the advantages of VHSR images that favor GEOBIA have been enumerated, the rich information in VHSR images leads to higher intraclass variation and lower interclass differences. Specifically, agricultural landscapes, especially smallholder farms, are covered by complex and diverse land use categories; hence, the spectral, shape, and texture features of these landscapes on VHSR images change over time and space, leading to greater internal variability [
12,
23]. To effectively address this problem, classification methods with good predictive ability and robustness should be considered in VHSR image analysis [
24].
Parallel to the advancements in VHSR satellite imagery, the development, and application of machine learning algorithms (MLAs) for performing image classification has gradually become a focus in the RS field [
31]. MLAs have been increasingly used in RS-based crop mapping, due to their rapid learning and adaptation to nonlinearity [
29,
32,
33]. Obviously, a variety of crop classification models have been developed based on various MLAs, such as models based on decision trees, artificial neural networks, support vector machines (SVMs), and the
k-nearest neighbors algorithm (
k-NN) [
32,
34,
35]. However, these models generally rely on a single MLA-based classifier and are prone to overfitting with limited training data [
36,
37]. Especially in complex agricultural areas, the accuracy of crop mapping based on a single classifier is often limited [
32,
38,
39]. For instance, Zhang et al. [
5] built several models by implementing individual SVM classifiers on different image features to distinguish smallholder crop types from WorldView-2 (WV2) images, and the accuracy of these models was less than 80%. In addition, there is no overall optimal MLA for crop classification modeling, and the best MLA generally depends on the objective of the classification task, the details of the problem, and the data structure used [
31]. Currently, increasing attention is being paid to further improving the existing applications of individual MLAs using ensemble learning (EL) techniques [
37,
39].
The use of EL technology to improve the accuracy of smallholder crop mapping with VHSR images remains to be further explored. EL is defined as a collection of methods that improves prediction performance by training multiple classifiers and summarizing their output [
40]. Empirically, EL methods tend to perform better than a single classifier in most cases unless the individual classifiers involved in the ensemble fail to provide sufficient diversity of generalization patterns [
36,
41]. Various EL methods have been proposed, which can generally be divided into two categories: Homogeneous and heterogeneous ensemble methods [
40]. The former combine multiple instances of the same MLA trained on several random subsets of the original training dataset; an example is bagging methods [
42]. The latter combine several different individual MLAs trained on the same dataset; an example is stacking methods [
43]. In practice, the random forest (RF) algorithm based on decision trees, as a typical example of the ready-made bagging method, has been widely used in crop mapping [
12,
21,
33]. However, bagging methods based on other MLAs have rarely been compared and tested for crop classification. In recent years, stacking methods have gradually been used to improve grain yield prediction and land use and land cover (LULC) classification. For example, Feng et al. [
37] improved the accuracy of yield prediction in the United States using a stacking model combining RF, SVM, and
k-NN classifiers. Man et al. [
39] improved the land cover classification in frequently cloud-covered areas by constructing an ensemble model combining five individual classifiers. In summary, although EL methods have been increasingly used to improve LULC mapping, studies focusing on their potential to improve the fine-scale mapping of smallholder crops from VHSR images have been rare.
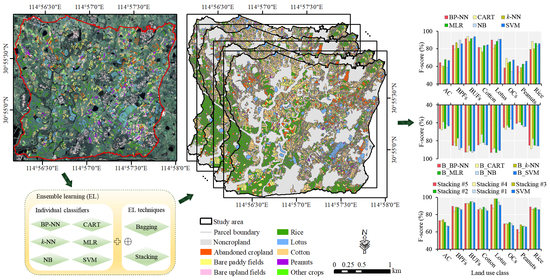
Therefore, this study aims to develop an ensemble machine learning-based framework to improve parcel-level crop mapping on smallholder farms from VHSR images. Our experiments were conducted on WV2 images from a typical smallholder agricultural area in central China. Six widely used classifiers with different basic ideas, namely, multinomial logistic regression (MLR), naive Bayes (NB) classifier, classification and regression tree (CART), backpropagation neural networks (BP-NN), k-NN, and SVM classifiers, were considered base classifiers. Bagging and stacking are typical examples of homogeneous and heterogeneous EL techniques, respectively, and therefore, were chosen to combine base classifiers. The specific objectives are to: (1) Explore how to build an appropriate ensemble model to achieve fine mapping of smallholder crops; (2) assess and compare the effects of bagging and stacking in improving the performance of individual classifiers; and (3) analyze the impact of parcel-level spatial information (e.g., textural and geometric features) extracted from VHSR images on the performance of the EL-based model.
6. Conclusions
Timely and accurate mapping of smallholder crops at the parcel level is essential for predicting grain yields and formulating area-based subsidies. In this study, an ensemble machine-learning-based framework was presented to improve the accuracy of parcel-level smallholder crop mapping from VHSR images. Several ensemble models were built by applying bagging and stacking approaches separately on six widely used individual classifiers. The comparative experiments showed that the stacking approach was superior to the bagging approach in improving the mapping accuracy of smallholder crops based on individual classifiers. The bagging models enhanced the performance of classifiers with tree structures or neural networks (e.g., CART and BP-NN), but these improvements were limited in that they narrowed only the accuracy gap between the ‘bad’ classifiers and the ‘good’ classifiers. The stacking models tended to perform better, and the Stacking #2 model, which uses the SVM as a meta-classifier to integrate the three best-performing individual classifiers (i.e., SVM, MLR, and CART), performed the best among all of the built models and improved the classwise accuracy of almost all of the land use types. This ensemble approach does not require additional costly sampling and specialized equipment, and improvements in mapping accuracy are clearly valuable for the delicacy management of smallholder crops. In addition, we also proved that using the spatial features of image objects can improve the accuracy of parcel-level smallholder crop mapping. In summary, the proposed framework shows the great potential of combining ensemble learning technology with VHSR images for accurate mapping of smallholder crops, which could facilitate the development of parcel-level crop identification systems in countries dominated by smallholder agriculture.
Although these experiments focused on smallholder farms in central China, the methodological framework presented in this study could easily be applied to other similarly complex and heterogeneous agricultural areas. In the future, methods for mapping intercropping and mixed-cropping patterns need to be developed to improve the classification accuracy of smallholder crops. In addition, independent of the classifier integration, image composition accounting for phenology to support dynamic mapping of smallholder crops needs further research.