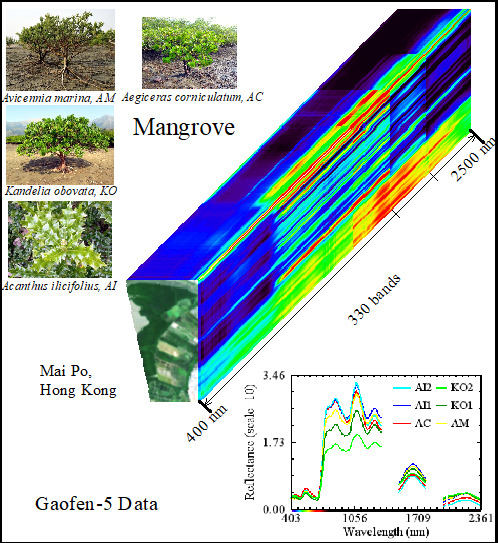
GF-5 Hyperspectral Data for Species Mapping of Mangrove in Mai Po, Hong Kong
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Dataset and Preprocessing
2.2.1. Hyperspectral Data and Preprocessing
2.2.2. Field Survey and Sample Data
2.3. Methods
2.3.1. Generation of Simulated Hyperion Data
2.3.2. Mangrove Species Classification with Machine Learning Methods
2.3.3. Accuracy Assessment
3. Results
3.1. Comparison of Simulated Hyprion and GF-5
3.2. Mangrove Species Mapping with Accuracy Assessment
4. Discussion
4.1. Quality Assessment of Simulated Hyprion
4.2. Classifier Selection
4.3. Contribution of the Increase in Spectral Resolution in VNIR
4.4. Limitation of the Study
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| N.O. | VNIR/SWIR | Band Number in GF-5 | Wavelength (nm) | Note* | N.O. | VNIR/SWIR | Band Number in GF-5 | Wavelength (nm) | Note* |
|---|---|---|---|---|---|---|---|---|---|
| 1 | VNIR | 4 | 402.96 | 2 | 126 | VNIR | 134 | 959.07 | 2 |
| 2 | VNIR | 5 | 407.24 | 2 | 127 | VNIR | 135 | 963.35 | 1 |
| 3 | VNIR | 6 | 411.52 | 2 | 128 | VNIR | 136 | 967.63 | 2 |
| 4 | VNIR | 7 | 415.80 | 2 | 129 | VNIR | 137 | 971.91 | 1 |
| 5 | VNIR | 8 | 420.08 | 2 | 130 | VNIR | 138 | 976.18 | 2 |
| 6 | VNIR | 9 | 424.36 | 2 | 131 | VNIR | 139 | 980.46 | 2 |
| 7 | VNIR | 10 | 428.64 | 2 | 132 | VNIR | 140 | 984.74 | 1 |
| 8 | VNIR | 11 | 432.91 | 2 | 133 | VNIR | 141 | 989.02 | 2 |
| 9 | VNIR | 12 | 437.19 | 2 | 134 | VNIR | 142 | 993.30 | 1 |
| 10 | VNIR | 13 | 441.47 | 2 | 135 | VNIR | 143 | 997.76 | 2 |
| 11 | VNIR | 14 | 445.75 | 2 | 136 | VNIR | 144 | 1002.22 | 1 |
| 12 | VNIR | 15 | 450.03 | 2 | 137 | VNIR | 145 | 1006.68 | 2 |
| 13 | VNIR | 16 | 454.31 | 2 | 138 | VNIR | 146 | 1011.14 | 2 |
| 14 | VNIR | 17 | 458.59 | 2 | 139 | VNIR | 147 | 1015.60 | 1 |
| 15 | VNIR | 18 | 462.87 | 2 | 140 | VNIR | 148 | 1020.06 | 2 |
| 16 | VNIR | 19 | 467.15 | 2 | 141 | VNIR | 149 | 1024.52 | 1 |
| 17 | VNIR | 20 | 471.42 | 2 | 142 | VNIR | 150 | 1028.98 | 2 |
| 18 | VNIR | 21 | 475.70 | 2 | 143 | SWIR | 5 | 1038.28 | 1 |
| 19 | VNIR | 22 | 479.98 | 1 | 144 | SWIR | 6 | 1046.71 | 1 |
| 20 | VNIR | 23 | 484.26 | 2 | 145 | SWIR | 7 | 1055.13 | 1 |
| 21 | VNIR | 24 | 488.54 | 1 | 146 | SWIR | 8 | 1063.56 | 1 |
| 22 | VNIR | 25 | 492.82 | 2 | 147 | SWIR | 9 | 1071.99 | 1 |
| 23 | VNIR | 26 | 497.10 | 2 | 148 | SWIR | 10 | 1080.42 | 3 |
| 24 | VNIR | 27 | 501.38 | 1 | 149 | SWIR | 11 | 1088.84 | 1 |
| 25 | VNIR | 28 | 505.66 | 2 | 150 | SWIR | 12 | 1097.27 | 1 |
| 26 | VNIR | 29 | 509.94 | 1 | 151 | SWIR | 13 | 1105.70 | 1 |
| 27 | VNIR | 30 | 514.22 | 2 | 152 | SWIR | 14 | 1114.12 | 1 |
| 28 | VNIR | 31 | 518.49 | 1 | 153 | SWIR | 20 | 1164.69 | 1 |
| 29 | VNIR | 32 | 522.77 | 2 | 154 | SWIR | 21 | 1173.12 | 1 |
| 30 | VNIR | 33 | 527.05 | 2 | 155 | SWIR | 22 | 1181.54 | 1 |
| 31 | VNIR | 34 | 531.33 | 1 | 156 | SWIR | 23 | 1189.97 | 3 |
| 32 | VNIR | 35 | 535.61 | 2 | 157 | SWIR | 24 | 1198.40 | 1 |
| 33 | VNIR | 36 | 539.94 | 1 | 158 | SWIR | 25 | 1206.60 | 1 |
| 34 | VNIR | 37 | 544.20 | 2 | 159 | SWIR | 26 | 1215.00 | 1 |
| 35 | VNIR | 38 | 548.47 | 1 | 160 | SWIR | 27 | 1223.40 | 1 |
| 36 | VNIR | 39 | 552.71 | 2 | 161 | SWIR | 28 | 1232.14 | 1 |
| 37 | VNIR | 40 | 556.97 | 2 | 162 | SWIR | 29 | 1240.56 | 3 |
| 38 | VNIR | 41 | 561.26 | 1 | 163 | SWIR | 30 | 1249.01 | 1 |
| 39 | VNIR | 42 | 565.55 | 2 | 164 | SWIR | 31 | 1257.46 | 1 |
| 40 | VNIR | 43 | 569.83 | 1 | 165 | SWIR | 32 | 1265.90 | 1 |
| 41 | VNIR | 44 | 574.12 | 2 | 166 | SWIR | 33 | 1274.35 | 1 |
| 42 | VNIR | 45 | 578.40 | 2 | 167 | SWIR | 34 | 1282.80 | 1 |
| 43 | VNIR | 46 | 582.69 | 1 | 168 | SWIR | 35 | 1291.25 | 3 |
| 44 | VNIR | 47 | 586.97 | 2 | 169 | SWIR | 36 | 1299.70 | 1 |
| 45 | VNIR | 48 | 591.26 | 1 | 170 | SWIR | 37 | 1308.14 | 1 |
| 46 | VNIR | 49 | 595.54 | 2 | 171 | SWIR | 38 | 1316.59 | 1 |
| 47 | VNIR | 50 | 599.83 | 1 | 172 | SWIR | 39 | 1325.04 | 1 |
| 48 | VNIR | 51 | 604.11 | 2 | 173 | SWIR | 61 | 1510.89 | 1 |
| 49 | VNIR | 52 | 608.40 | 2 | 174 | SWIR | 62 | 1519.34 | 1 |
| 50 | VNIR | 53 | 612.69 | 1 | 175 | SWIR | 63 | 1527.79 | 1 |
| 51 | VNIR | 54 | 616.97 | 2 | 176 | SWIR | 64 | 1536.23 | 1 |
| 52 | VNIR | 55 | 621.26 | 1 | 177 | SWIR | 65 | 1544.68 | 1 |
| 53 | VNIR | 56 | 625.54 | 2 | 178 | SWIR | 66 | 1553.13 | 3 |
| 54 | VNIR | 57 | 629.83 | 1 | 179 | SWIR | 67 | 1560.73 | 1 |
| 55 | VNIR | 58 | 634.11 | 2 | 180 | SWIR | 68 | 1569.03 | 1 |
| 56 | VNIR | 59 | 638.40 | 2 | 181 | SWIR | 69 | 1577.41 | 1 |
| 57 | VNIR | 60 | 642.68 | 1 | 182 | SWIR | 70 | 1586.11 | 1 |
| 58 | VNIR | 61 | 646.88 | 2 | 183 | SWIR | 71 | 1594.76 | 3 |
| 59 | VNIR | 62 | 651.11 | 1 | 184 | SWIR | 72 | 1603.18 | 1 |
| 60 | VNIR | 63 | 655.35 | 2 | 185 | SWIR | 73 | 1611.59 | 1 |
| 61 | VNIR | 64 | 659.63 | 2 | 186 | SWIR | 74 | 1620.01 | 1 |
| 62 | VNIR | 65 | 663.91 | 1 | 187 | SWIR | 75 | 1628.43 | 1 |
| 63 | VNIR | 66 | 668.24 | 2 | 188 | SWIR | 76 | 1636.85 | 1 |
| 64 | VNIR | 67 | 672.60 | 1 | 189 | SWIR | 77 | 1645.27 | 3 |
| 65 | VNIR | 68 | 676.90 | 2 | 190 | SWIR | 78 | 1653.69 | 1 |
| 66 | VNIR | 69 | 681.19 | 1 | 191 | SWIR | 79 | 1662.11 | 1 |
| 67 | VNIR | 70 | 685.42 | 2 | 192 | SWIR | 80 | 1670.53 | 1 |
| 68 | VNIR | 71 | 689.68 | 2 | 193 | SWIR | 81 | 1678.95 | 1 |
| 69 | VNIR | 72 | 693.95 | 1 | 194 | SWIR | 82 | 1687.37 | 1 |
| 70 | VNIR | 73 | 698.17 | 2 | 195 | SWIR | 83 | 1695.79 | 3 |
| 71 | VNIR | 74 | 702.39 | 1 | 196 | SWIR | 84 | 1704.21 | 1 |
| 72 | VNIR | 75 | 706.67 | 2 | 197 | SWIR | 85 | 1712.63 | 1 |
| 73 | VNIR | 76 | 710.95 | 2 | 198 | SWIR | 86 | 1721.05 | 1 |
| 74 | VNIR | 77 | 715.23 | 1 | 199 | SWIR | 87 | 1729.47 | 1 |
| 75 | VNIR | 78 | 719.51 | 2 | 200 | SWIR | 88 | 1737.88 | 1 |
| 76 | VNIR | 79 | 723.79 | 1 | 201 | SWIR | 89 | 1746.30 | 3 |
| 77 | VNIR | 80 | 728.06 | 2 | 202 | SWIR | 90 | 1754.72 | 1 |
| 78 | VNIR | 81 | 732.34 | 1 | 203 | SWIR | 91 | 1763.14 | 1 |
| 79 | VNIR | 82 | 736.62 | 2 | 204 | SWIR | 92 | 1771.56 | 1 |
| 80 | VNIR | 83 | 740.90 | 2 | 205 | SWIR | 93 | 1779.98 | 1 |
| 81 | VNIR | 84 | 745.17 | 1 | 206 | SWIR | 94 | 1788.40 | 1 |
| 82 | VNIR | 85 | 749.45 | 2 | 207 | SWIR | 116 | 1973.63 | 3 |
| 83 | VNIR | 86 | 753.73 | 1 | 208 | SWIR | 117 | 1982.05 | 1 |
| 84 | VNIR | 87 | 758.01 | 2 | 209 | SWIR | 118 | 1990.47 | 1 |
| 85 | VNIR | 88 | 762.29 | 1 | 210 | SWIR | 122 | 2024.14 | 1 |
| 86 | VNIR | 89 | 766.57 | 2 | 211 | SWIR | 123 | 2032.56 | 1 |
| 87 | VNIR | 90 | 770.84 | 2 | 212 | SWIR | 124 | 2040.98 | 1 |
| 88 | VNIR | 91 | 775.12 | 1 | 213 | SWIR | 125 | 2049.40 | 1 |
| 89 | VNIR | 92 | 779.40 | 2 | 214 | SWIR | 126 | 2057.82 | 3 |
| 90 | VNIR | 93 | 783.68 | 1 | 215 | SWIR | 127 | 2066.24 | 1 |
| 91 | VNIR | 94 | 787.96 | 2 | 216 | SWIR | 128 | 2074.66 | 1 |
| 92 | VNIR | 95 | 792.23 | 2 | 217 | SWIR | 129 | 2083.08 | 1 |
| 93 | VNIR | 96 | 796.51 | 1 | 218 | SWIR | 130 | 2091.50 | 1 |
| 94 | VNIR | 97 | 800.79 | 2 | 219 | SWIR | 131 | 2099.92 | 1 |
| 95 | VNIR | 98 | 805.07 | 1 | 220 | SWIR | 132 | 2108.34 | 3 |
| 96 | VNIR | 99 | 809.34 | 2 | 221 | SWIR | 133 | 2116.77 | 1 |
| 97 | VNIR | 100 | 813.62 | 1 | 222 | SWIR | 134 | 2125.21 | 1 |
| 98 | VNIR | 101 | 817.90 | 2 | 223 | SWIR | 135 | 2134.10 | 1 |
| 99 | VNIR | 102 | 822.18 | 2 | 224 | SWIR | 136 | 2142.11 | 1 |
| 100 | VNIR | 103 | 826.46 | 1 | 225 | SWIR | 137 | 2150.68 | 1 |
| 101 | VNIR | 104 | 830.73 | 2 | 226 | SWIR | 138 | 2159.11 | 3 |
| 102 | VNIR | 105 | 835.01 | 1 | 227 | SWIR | 139 | 2167.53 | 1 |
| 103 | VNIR | 106 | 839.29 | 2 | 228 | SWIR | 140 | 2175.96 | 1 |
| 104 | VNIR | 107 | 843.57 | 1 | 229 | SWIR | 141 | 2184.39 | 1 |
| 105 | VNIR | 108 | 847.85 | 2 | 230 | SWIR | 142 | 2192.81 | 1 |
| 106 | VNIR | 109 | 852.12 | 2 | 231 | SWIR | 143 | 2201.24 | 1 |
| 107 | VNIR | 110 | 856.40 | 1 | 232 | SWIR | 144 | 2209.67 | 3 |
| 108 | VNIR | 111 | 860.68 | 2 | 233 | SWIR | 145 | 2218.10 | 1 |
| 109 | VNIR | 112 | 864.96 | 1 | 234 | SWIR | 146 | 2226.52 | 1 |
| 110 | VNIR | 113 | 869.23 | 2 | 235 | SWIR | 147 | 2234.95 | 1 |
| 111 | VNIR | 114 | 873.51 | 2 | 236 | SWIR | 148 | 2243.38 | 1 |
| 112 | VNIR | 115 | 877.79 | 1 | 237 | SWIR | 149 | 2251.81 | 1 |
| 113 | VNIR | 116 | 882.07 | 2 | 238 | SWIR | 150 | 2260.23 | 3 |
| 114 | VNIR | 117 | 886.35 | 1 | 239 | SWIR | 151 | 2268.66 | 1 |
| 115 | VNIR | 118 | 890.63 | 2 | 240 | SWIR | 152 | 2277.09 | 1 |
| 116 | VNIR | 119 | 894.90 | 1 | 241 | SWIR | 153 | 2285.51 | 1 |
| 117 | VNIR | 120 | 899.18 | 2 | 242 | SWIR | 154 | 2293.94 | 1 |
| 118 | VNIR | 121 | 903.46 | 2 | 243 | SWIR | 155 | 2302.37 | 1 |
| 119 | VNIR | 122 | 907.74 | 1 | 244 | SWIR | 156 | 2310.80 | 3 |
| 120 | VNIR | 123 | 912.02 | 2 | 245 | SWIR | 157 | 2319.22 | 1 |
| 121 | VNIR | 124 | 916.29 | 1 | 246 | SWIR | 158 | 2327.65 | 1 |
| 122 | VNIR | 125 | 920.57 | 2 | 247 | SWIR | 159 | 2336.08 | 1 |
| 123 | VNIR | 126 | 924.85 | 1 | 248 | SWIR | 160 | 2344.51 | 1 |
| 124 | VNIR | 127 | 929.13 | 2 | 249 | SWIR | 161 | 2352.93 | 1 |
| 125 | VNIR | 133 | 954.79 | 2 | 250 | SWIR | 162 | 2361.36 | 1 |
References
- Wang, L.; Jia, M.; Yin, D.; Tian, J. A review of remote sensing for mangrove forests: 1956–2018. Remote Sens. Environ. 2019, 231, 111223. [Google Scholar] [CrossRef]
- Giri, C.; Ochieng, E.; Tieszen, L.L.; Zhu, Z.; Singh, A.; Loveland, T.; Masek, J.; Duke, N. Status and distribution of mangrove forests of the world using earth observation satellite data. Glob. Ecol. Biogeogr. 2011, 20, 154–159. [Google Scholar] [CrossRef]
- Hamilton, S.E.; Casey, D. Creation of a high spatio-temporal resolution global database of continuous mangrove forest cover for the 21st century (CGMFC-21). Glob. Ecol. Biogeogr. 2016, 25, 729–738. [Google Scholar] [CrossRef]
- Duke, N.C.; Meynecke, J.O.; Dittmann, S.; Ellison, A.M.; Anger, K.; Berger, U.; Cannicci, S.; Diele, K.; Ewel, K.C.; Field, C.D.; et al. A World Without Mangroves? Science 2007, 317, 41. [Google Scholar] [CrossRef] [Green Version]
- Kovacs, J.M.; Wang, J.; Flores-Verdugo, F. Mapping mangrove leaf area index at the species level using IKONOS and LAI-2000 sensors for the Agua Brava Lagoon, Mexican Pacific. Estuar. Coast. Shelf Sci. 2005, 62, 377–384. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, H.S.; Lin, H.; Fang, C.Y. Textural-Spectral Feature-Based Species Classification of Mangroves in Mai Po Nature Reserve from Worldview-3 Imagery. Remote Sens. 2016, 8, 24. [Google Scholar] [CrossRef] [Green Version]
- Fatoyinbo, T.; Feliciano, E.A.; Lagomasino, D.; Lee, S.K.; Trettin, C. Estimating mangrove aboveground biomass from airborne LiDAR data: A case study from the Zambezi River delta. Environ. Res. Lett. 2018, 13. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Zhang, H.; Lin, G.; Lin, H.; Tang, D. Zonation and Directional Dynamics of Mangrove Forests Derived from Time-Series Satellite Imagery in Mai Po, Hong Kong. Sustainability 2018, 10, 1913. [Google Scholar] [CrossRef] [Green Version]
- Pham, T.D.; Yokoya, N.; Bui, D.T.; Yoshino, K.; Friess, D.A. Remote Sensing Approaches for Monitoring Mangrove Species, Structure, and Biomass: Opportunities and Challenges. Remote Sens. 2019, 11, 230. [Google Scholar] [CrossRef] [Green Version]
- Yin, D.M.; Wang, L. Individual mangrove tree measurement using UAV-based LiDAR data: Possibilities and challenges. Remote Sens. Environ. 2019, 223, 34–49. [Google Scholar] [CrossRef]
- Pham, T.D.; Bui, D.T.; Yoshino, K.; Le, N.N. Optimized rule-based logistic model tree algorithm for mapping mangrove species using ALOS PALSAR imagery and GIS in the tropical region. Environ. Earth Sci. 2018, 77. [Google Scholar] [CrossRef]
- Zhang, H.; Li, J.; Wang, T.; Lin, H.; Zheng, Z.; Li, Y.; Lu, Y. A manifold learning approach to urban land cover classification with optical and radar data. Landsc. Urban Plan. 2018, 172, 11–24. [Google Scholar] [CrossRef]
- Wang, L.; Sousa, W.P.; Gong, P. Integration of object-based and pixel-based classification for mapping mangroves with IKONOS imagery. Int. J. Remote Sens. 2004, 25, 5655–5668. [Google Scholar] [CrossRef]
- Jia, M.M.; Zhang, Y.Z.; Wang, Z.M.; Song, K.S.; Ren, C.Y. Mapping the distribution of mangrove species in the Core Zone of Mai Po Marshes Nature Reserve, Hong Kong, using hyperspectral data and high-resolution data. Int. J. Appl. Earth Observ. Geoinf. 2014, 33, 226–231. [Google Scholar] [CrossRef]
- Neukermans, G.; Dahdouh-Guebas, F.; Kairo, J.G.; Koedam, N. Mangrove species and stand mapping in GAzi bay (Kenya) using Quickbird satellite imagery. J. Spat. Sci. 2008, 53, 75–86. [Google Scholar] [CrossRef]
- Wang, D.Z.; Wan, B.; Qiu, P.H.; Su, Y.J.; Guo, Q.H.; Wu, X.C. Artificial Mangrove Species Mapping Using Pleiades-1: An Evaluation of Pixel-Based and Object-Based Classifications with Selected Machine Learning Algorithms. Remote Sens. 2018, 10, 294. [Google Scholar] [CrossRef] [Green Version]
- Viennois, G.; Proisy, C.; Feret, J.B.; Prosperi, J.; Sidik, F.; Suhardjono; Rahmania, R.; Longepe, N.; Germain, O.; Gaspar, P. Multitemporal Analysis of High-Spatial-Resolution Optical Satellite Imagery for Mangrove Species Mapping in Bali, Indonesia. IEEE J. Sel. Top. Appl. Earth Observ. Remote Sens. 2016, 9, 3680–3686. [Google Scholar] [CrossRef]
- Wan, L.; Zhang, H.; Wang, T.; Li, G.; Lin, H. Mangrove Species Discrimination from Very High Resolution Imagery Using Gaussian Markov Random Field Model. Wetlands 2018, 38, 861–874. [Google Scholar] [CrossRef]
- Kamal, M.; Phinn, S.; Johansen, K. Object-Based Approach for Multi-Scale Mangrove Composition Mapping Using Multi-Resolution Image Datasets. Remote Sens. 2015, 7, 4753–4783. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Sousa, W.P.; Gong, P.; Biging, G.S. Comparison of IKONOS and QuickBird images for mapping mangrove species on the Caribbean coast of Panama. Remote Sens. Environ. 2004, 91, 432–440. [Google Scholar] [CrossRef]
- Vaiphasa, C.; Ongsomwang, S.; Vaiphasa, T.; Skidmore, A.K. Tropical mangrove species discrimination using hyperspectral data: A laboratory study. Estuar. Coast. Shelf Sci. 2005, 65, 371–379. [Google Scholar] [CrossRef]
- Wang, L.; Sousa, W.P. Distinguishing mangrove species with laboratory measurements of hyperspectral leaf reflectance. Int. J. Remote Sens. 2009, 30, 1267–1281. [Google Scholar] [CrossRef]
- Manjunath, K.R.; Kumar, T.; Kundu, N.; Panigrahy, S. Discrimination of mangrove species and mudflat classes using in situ hyperspectral data: A case study of Indian Sundarbans. Gisci. Remote Sens. 2013, 50, 400–417. [Google Scholar] [CrossRef]
- Zhang, C.H.; Kovacs, J.M.; Liu, Y.L.; Flores-Verdugo, F.; Flores-de-Santiago, F. Separating Mangrove Species and Conditions Using Laboratory Hyperspectral Data: A Case Study of a Degraded Mangrove Forest of the Mexican Pacific. Remote Sens. 2014, 6, 11673–11688. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.J.; Liu, K.; Liu, L.; Zhu, Y.H.; Li, J.; He, Z. Identifying Mangrove Species Using Field Close-Range Snapshot Hyperspectral Imaging and Machine-Learning Techniques. Remote Sens. 2018, 10, 2047. [Google Scholar] [CrossRef] [Green Version]
- Held, A.; Ticehurst, C.; Lymburner, L.; Williams, N. High resolution mapping of tropical mangrove ecosystems using hyperspectral and radar remote sensing. Int. J. Remote Sens. 2003, 24, 2739–2759. [Google Scholar] [CrossRef]
- Chaube, N.R.; Lele, N.; Misra, A.; Murthy, T.V.R.; Manna, S.; Hazra, S.; Panda, M.; Samal, R.N. Mangrove species discrimination and health assessment using AVIRIS-NG hyperspectral data. Curr. Sci. 2019, 116, 1136–1142. [Google Scholar] [CrossRef]
- Wong, K.K. Mangrove Species Mapping and Leaf Area Index Modeling Using Optical and Microwave Remote Sensing Technologies in Hong Kong. Phd, The Chinese University of Hong Kong, Hong Kong, 2012.
- Koedsin, W.; Vaiphasa, C. Discrimination of Tropical Mangroves at the Species Level with EO-1 Hyperion Data. Remote Sens. 2013, 5, 3562. [Google Scholar] [CrossRef] [Green Version]
- Krebs, G. Gaofen 5 (GF 5). Available online: https://space.skyrocket.de/doc_sdat/gf-5.htm (accessed on 23 December 2019).
- Liu, Y.; Sun, D.; Hu, X.; Ye, X.; Li, Y.; Liu, S.; Cao, K.; Chai, M.; Zhou, W.; Zhang, J.; et al. The Advanced Hyperspectral Imager: Aboard China’s GaoFen-5 Satellite. IEEE Geosci. Remote Sens. Mag. 2019, 7, 23–32. [Google Scholar] [CrossRef]
- Kuenzer, C.; Bluemel, A.; Gebhardt, S.; Quoc, T.V.; Dech, S. Remote Sensing of Mangrove Ecosystems: A Review. Remote Sens. 2011, 3, 878–928. [Google Scholar] [CrossRef] [Green Version]
- Wong, F.K.; Fung, T. Combining EO-1 Hyperion and Envisat ASAR data for mangrove species classification in Mai Po Ramsar Site, Hong Kong. Int. J. Remote Sens. 2014, 35, 7828–7856. [Google Scholar] [CrossRef]
- Tong, Q.; Zhang, B.; Zhang, L. Current progress of hyperspectral remote sensing in China. J. Remote Sens. 2016, 20, 689–707. [Google Scholar] [CrossRef]
- Zhang, B. Advancement of hyperspectral image processing and information extraction. J. Remote Sens. 2016, 20, 1062–1090. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.Z.; Zhang, H.S.; Lin, H. Improving the impervious surface estimation with combined use of optical and SAR remote sensing images. Remote Sens. Environ. 2014, 141, 155–167. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Pal, M.; Foody, G.M. Feature Selection for Classification of Hyperspectral Data by SVM. IEEE Trans. Geosci. Remote Sens. 2010, 48, 2297–2307. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.-C.; Lin, C.-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 1–27. [Google Scholar] [CrossRef]
- Hsu, C.-W.; Chang, C.-C.; Lin, C.-J. A Practical Guide to Support Vector Classification. Available online: https://www.csie.ntu.edu.tw/~cjlin/papers/guide/guide.pdf (accessed on 24 May 2019).
- Congalton, R.G. Assessing Landsat Classification Accuracy Using Discrete Multivariate Analysis Statistical Techniques. Photogramm. Eng. Remote Sens. 1983, 49, 1671–1678. [Google Scholar]
- Pham, L.T.H.; Brabyn, L. Monitoring mangrove biomass change in Vietnam using SPOT images and an object-based approach combined with machine learning algorithms. Isprs J. Photogramm. Remote Sens. 2017, 128, 86–97. [Google Scholar] [CrossRef]





| Data | Band | Spectral Range (nm) | Spectral Resolution (nm) | Spatial Resolution (m) | Bands |
|---|---|---|---|---|---|
| GF-5 | VNIR | 390.324–1029.18 | 5 | 30 | 150 |
| SWIR | 1004.77–2513.25 | 10 | 30 | 180 | |
| Hyperion | VNIR | 355.59–1057.68 | 10 | 30 | 70 |
| SWIR | 851.9–2577.08 | 10 | 30 | 172 |
| Data Quality | Band N.O. | Bands |
|---|---|---|
| No Calibration | SWIR: 43–50, 96–112 | 25 |
| Strip contamination | VNIR: 1–3; SWIR: 40–42, 51–60, 95, 113–115, 119–121, 163–180 | 41 |
| Absorption* | VNIR: 128–132; SWIR: 15–19, 43–57, 96–115 | 45 |
| Spectral Overlap in SWIR | SWIR: 1–4 | 4 |
| Species | Species Code | Description [6,33] | Sample Number (pixel) |
|---|---|---|---|
| Kandelia obovate | KO1 | locate at the landward side, tend to be high | 87 |
| KO2 | locate at the seaward side; tend to be low | 26 | |
| Avicennia marina | AM | locates in the central part of the Mai Po | 70 |
| Acanthus ilicifolius | AI1 | locates at the landward side; tend to be with serrated leaves | 49 |
| AI2 | locates at the seaward side; tend to be with smooth leaves | 42 | |
| Aegiceras corniculatum | AC | usually found as undergrowth | 19 |
| Data | Bandwidth (nm) | Bands |
|---|---|---|
| Landsat 81 | 430-450, 450-510, 530-590, 630-670, 850-880, 1570-1650, 2110-2290 | 7 |
| Simulated Hyperion (SH) | The value of 1 in ‘Note’ field (See Appendix A) | 144 |
| SH + extra bands in VNIR of GF-5 (SH-GF5VN) | The value of 1 and 2 in ‘Note’ field (See Appendix A) | 234 |
| SH + extra bands in SWIR of GF-5 (SH-GF5SW) | The value of 1 and 3 in ‘Note’ field (See Appendix A) | 160 |
| GF5 | 403–929, 955–1029, 1038–1325, 1511–1788, 1974–1990, 2024–2361 | 250 |
| P-Value (<0.05) | SH, SH-GF5VN | SH, SH-GF5SW | SH, GF-5 |
|---|---|---|---|
| Random Forests | 0.76283 | 0.91744 | 0.61711 |
| SVM | 0.37322 | 0.38491 | 0.99998 |
| Data | KO2 | KO1 | AM | AI2 | AI1 | AC | Total | User Acc (%) | |
|---|---|---|---|---|---|---|---|---|---|
| Landsat 8 | unclassified | 0.5 | 0.3 | 0 | 0 | 0 | 0 | 0.3 | |
| KO2 | 5.9 | 0.2 | 0 | 0 | 0 | 0 | 6.1 | 96.72 | |
| KO1 | 0 | 17.7 | 4.7 | 0.8 | 1.6 | 0.1 | 24.9 | 71.08 | |
| AM | 0 | 4.6 | 14.9 | 0.1 | 2.3 | 0 | 21.9 | 68.04 | |
| AI2 | 0.3 | 0.6 | 0 | 10 | 0 | 1 | 11.9 | 84.03 | |
| AI1 | 0 | 2.6 | 1.4 | 0 | 10.1 | 0 | 14.1 | 71.63 | |
| AC | 0 | 0 | 0 | 1 | 0 | 3.9 | 4.9 | 79.59 | |
| Total | 6.7 | 26 | 21 | 11.9 | 14 | 5 | 84.6 | ||
| Prod Acc (%) | 88.06 | 68.08 | 70.95 | 84.03 | 72.14 | 78.00 | |||
| SH | unclassified | 0 | 0.3 | 0 | 0 | 0 | 0 | 0.3 | |
| KO2 | 6.5 | 0.7 | 0 | 0 | 0 | 0 | 7.2 | 90.28 | |
| KO1 | 0.5 | 22.1 | 1.3 | 0 | 0.4 | 0 | 24.3 | 90.95 | |
| AM | 0 | 2.9 | 18.3 | 0 | 2.2 | 0 | 23.4 | 78.21 | |
| AI2 | 0 | 0 | 0 | 11.6 | 0 | 1.1 | 12.7 | 91.34 | |
| AI1 | 0 | 0 | 1.4 | 0 | 11.4 | 0 | 12.8 | 89.06 | |
| AC | 0 | 0 | 0 | 0.4 | 0 | 3.9 | 4.3 | 90.70 | |
| Total | 7 | 26 | 21 | 12 | 14 | 5 | 85 | ||
| Prod Acc (%) | 92.86 | 85.00 | 87.14 | 96.67 | 81.43 | 78.00 | |||
| SH-GF5VN | unclassified | 0 | 0.3 | 0 | 0 | 0 | 0 | 0.3 | |
| KO2 | 6.3 | 0.6 | 0 | 0 | 0 | 0 | 6.9 | 91.30 | |
| KO1 | 0.7 | 22.2 | 1.3 | 0 | 0.4 | 0 | 24.6 | 90.24 | |
| AM | 0 | 2.9 | 18 | 0 | 2.2 | 0 | 23.1 | 77.92 | |
| AI2 | 0 | 0 | 0 | 11.6 | 0 | 0.7 | 12.3 | 94.31 | |
| AI1 | 0 | 0 | 1.7 | 0 | 11.4 | 0.1 | 13.2 | 86.36 | |
| AC | 0 | 0 | 0 | 0.4 | 0 | 4.2 | 4.6 | 91.30 | |
| Total | 7 | 26 | 21 | 12 | 14 | 5 | 85 | ||
| Prod Acc (%) | 90.00 | 85.38 | 85.71 | 96.67 | 81.43 | 84.00 |
| Species | KO1 | AM | AI2 | AI1 | KO2 | AC |
|---|---|---|---|---|---|---|
| Area (ha) | 103.95±3.56 | 45.15±2.87 | 34.28±1.82 | 22.42±2.23 | 14.36±1.02 | 6.93±1.07 |
| Percentage (%) | 46 | 20 | 15 | 10 | 6 | 3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, L.; Lin, Y.; Zhang, H.; Wang, F.; Liu, M.; Lin, H. GF-5 Hyperspectral Data for Species Mapping of Mangrove in Mai Po, Hong Kong. Remote Sens. 2020, 12, 656. https://doi.org/10.3390/rs12040656
Wan L, Lin Y, Zhang H, Wang F, Liu M, Lin H. GF-5 Hyperspectral Data for Species Mapping of Mangrove in Mai Po, Hong Kong. Remote Sensing. 2020; 12(4):656. https://doi.org/10.3390/rs12040656
Chicago/Turabian StyleWan, Luoma, Yinyi Lin, Hongsheng Zhang, Feng Wang, Mingfeng Liu, and Hui Lin. 2020. "GF-5 Hyperspectral Data for Species Mapping of Mangrove in Mai Po, Hong Kong" Remote Sensing 12, no. 4: 656. https://doi.org/10.3390/rs12040656