1. Introduction
It has been demonstrated that close remote sensing approaches in combination with appropriate experimental designs and data integration can increase accuracy, precision, and throughput of on-field phenotyping experiments while also reducing cost and labor requirements [
1]. These high throughput phenotyping approaches can also be implemented in crop breeding to determine architectural/morphological and physiological traits to aid early detection of desirable genotypes [
2]. In these areas, the use of unmanned aerial vehicles (UAVs) is increasing steadily, as UAVs are suitable for use in complex field environments (e.g., muddy soil), are non-invasive, flexible in use (e.g., not restricted in use to particular field plots), and are available at a low cost [
3]. UAV flights can be scheduled in function of the needs of the breeding program or experiment, using combinations of sensors that are suitable to investigate the traits of interest. In one of the most straightforward applications, canopy cover (CC) and canopy height (CH) data can be derived from images obtained using an RGB camera mounted on an UAV [
4]. Canopy cover, defined as the ratio of the projected plant area to the total area considered, goes through changes from crop establishment to senescence. CC yields information regarding seedling emergence, early vigor, or senescence [
5] and is therefore an important criterion in a breeding context [
6]. Furthermore, CC data can be used to estimate canopy photosynthesis and transpiration [
7] or to evaluate crop responses to abiotic stresses (e.g., drought stress; [
8]). Canopy height (CH) is also dynamic and linked to growth, crop development, and yield [
9,
10].
One of the main advantages of UAV-based imaging approaches is the high precision and throughput for data acquisition and processing in comparison to the manual ruler measurements or scoring systems that have been traditionally used to analyze crop growth and development. By performing UAV flights throughout the entire growing season, accurate crop data can be obtained at a high spatio-temporal resolution, delivering dynamic information on crop growth and performance [
11]. For example, in Yu et al. (2016) [
12], data of five time points (using RGB and modified RGB-NIR imaging) were combined to classify soybean populations in different maturity groups and to estimate final yield. In Holman et al. (2016) [
13], a method for deriving crop height and growth rate from multi-temporal 3D digital surface models of winter wheat field trials was developed. In Pugh et al. (2018) [
14], UAV-based approaches to estimate height in sorghum and maize were determined and growth curves were fitted for different groups of breeding material. Similarly, in Chang et al. (2017) [
15] sigmoidal curves were fitted to UAV-derived multi-temporal height data of sorghum. Multi-temporal height data (four flights) have also been used in combination with clustering analysis methods to detect different growth patterns in maize varieties [
16].
These examples illustrate the value of multi-temporal data to describe crop growth and to predict specific parameters such as maturity, growth rate (in the above examples this was restricted to estimations considering two consecutive measurements) or yield. Up to now, however, little attention was paid to the biological interpretation of the growth curves built using UAV-based information. Nevertheless, the possibility for such interpretation represents one of the main advantages of this integrative approach. Multi-temporal data at high resolution can be of great value for sophisticated plant and crop modeling approaches. This is because crop models typically contain a large number of parameters that need to be estimated in an identifiable manner [
17] based on preferentially large quantities of objective experimental (field) data. Furthermore, mathematical models can be fitted to multi-temporal data to link growth patterns to relevant environmental and developmental clues for a precise description of crop performance from sowing to harvest. This might also enable derivation of ‘hidden’ variables as phenome descriptors that are useful in a breeding context (e.g., determinacy).
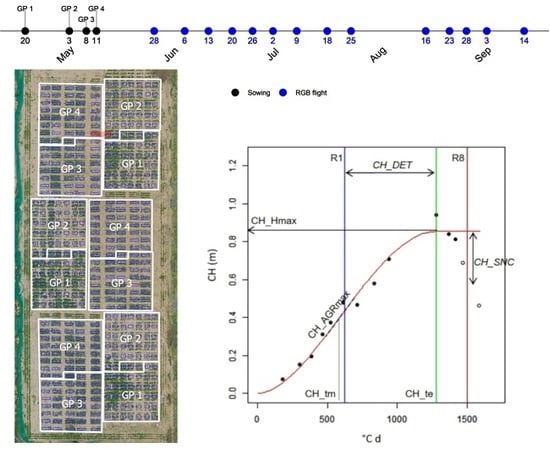
In this study, we demonstrate this concept using a soybean (Glycine max (L.) Merr.) experiment with 454 field plots comprising a diverse set of genotypes. More specifically, we fit mathematical models to multi-temporal data of CC and CH obtained for each plot and explore the possibilities of a global description of crop growth and development throughout the season in the context of breeding applications. First, CC and CH from each flight are estimated. Second, curves are fitted considering multi-temporal data per plot to extract biologically relevant information. Third, the correlation/complementarity of the model-based parameters obtained are evaluated. Fourth, parameters are examined for a possible relation to seed yield and maturity. Finally, an example to illustrate how the data could be used in a breeding context is developed to select interesting plots or to depict variability in the collection of genotypes evaluated.
3. Results
3.1. Model Fitting for Canopy Cover and Canopy Height
In an exploratory data analysis of CC and CH data, after discarding plots with emergence <50% or R2 < 0.7 in the estimation of R1 stage, different growth patterns per group were observed (data not shown), which was confirmed in the subsequent analysis.
Fitting of CC data using the Gompertz function resulted in 90.4% fits with an adjusted R
2 > 0.70. For the CH data to which the Beta model was fitted, 99.4% fits had an adjusted R
2 > 0.70. Combining the 0.7 limits for both fits, rendered 291 plots that fulfilled both criteria (89.8%), with a median CC_RsqA of 0.95 and CH_RsqA of 0.98 (
Figure 3).
The parameter values for CC are presented in
Table 2. For CC_tm (°C d) the median values ranged from 299 to 446; for CC_Cmax (%) the values ranged from 94.2 to 95.6. On average, CC_tm was lower for the later-sown groups GP3 and GP4 than for the groups GP1 and GP2 (
Figure 4A).
For CH, the parameter values are presented in
Table 3. For CH_tm (°C d) this resulted in a range from 547 to 735, for CH_Hmax (m) from 0.76 to 0.96 and for CH_te (°C d) from 901 to 1312.
To complete the results presented in
Figure 4;
Figure 5, we checked whether the differences between groups were significant. For CC_Cmax, the only pair with a significant difference was GP2-GP3. All pairwise comparisons were significant for CC_tm.
In the case of CH_Hmax only GP4 showed significant differences with the other three groups. All six pairwise combinations were significant for CH_tm. Almost the same results were found for CH_te— except GP1-GP2, which was not significantly different.
3.2. Extra Phenotypic Parameters Derived from the Fitted Curves
As models were fitted to the time series, we could derive phenotypic parameters that correspond to traits that are hard to observe or measure in the field such as growth rate, weed suppression, determinacy or senescence.
The median of the CC_AGRmax (% °C
−1 d
−1) ranged from 0.475 to 0.761 when different GPs were considered. Although the four groups differed in CC_AGRmax (
Figure 4C) almost all their plots reached high CC_Cmax > 90–95% (
Figure 4B) as can be expected because standard deviations are <3.1%. Based on the canopy cover growth curve, a value related to weed suppression (
Table 2 and
Figure 4D) was derived by estimating the percentage of canopy cover reached six weeks after emergence (CC_6W (%)). The median values varied between 65.8 and 94.6 for the four groups considered. Strikingly, a large spread of values in GP1 and GP2 was observed, indicating a slow canopy development for some of the genotypes contained in these groups. Thermal time values when 75% of canopy coverage was reached (CC_75PCT; °C d) ranged from 372 to 525, meaning that GP1 and GP2 needed more time (were slower) to close canopy. A group effect was also present here, with considerable variation found within each group (
Figure 4D,E).
The median values for CH_AGRmax (m °C
−1 d
−1) ranged from 0.00118 to 0.00155. The two groups with the highest CH_AGRmax were GP3 and GP4; they reached their maximum height earlier (lower CH_te), but the other two groups showed a higher CH_Hmax (
Figure 5B–D). The determinacy values are summarized in
Table 3 and
Figure 5E. This variable indicates how long the plants kept growing after reaching R1. The median CH_DET values ranged from 339 to 644 (°C d).
Figure 5E shows that GP3 and GP4 contained more determinate genotypes than GP1 and GP2, but again with a large variation.
Finally, based both on the canopy cover and the height fits, senescence was estimated (
Table 2;
Table 3 and
Figure 4F and
Figure 5F). These senescence parameters describe the ratio of decrease for CC and CH between the maximum value derived from the model fitting and the average value of the two last flights. In other words, the higher the values, the more senescence was observed. For CC_SNC median values ranged from 0.074 to 0.815, and in the case of CH_SNC, the median values ranged from 0.245 to 0.639. GP4 showed the highest CC_SNC and CH_SNC values followed by GP3 (
Figure 4F and
Figure 5F). Taken together with results for determinacy, we can conclude that GP3 and GP4 contained more determinate genotypes that senesce earlier in the environment tested. The histograms of all parameters are available in the
Supplementary Materials (Figure S2 and Figure S3).
The analysis to identify significant differences between groups resulted in the following. In the case of CC_AGRmax, all six pairwise comparisons were significant, except GP1-GP4. For CC_6W and CC_SNC, all pairwise comparisons were significant. For CC_75PCT, all pairwise comparisons significant except for the comparison GP3-GP4. In the case of CH_AGRmax the two pairs that were not significantly different were GP1-GP2 and GP3-GP4. For CH_DET and CH_SNC all comparisons were significant except for GP1-GP2.
3.3. Relations between Phenotypic Parameters
To visualize the relations and complementarity among all phenotypic data, a PCA was carried out (
Figure 6) and pair-wise correlations were calculated (
Figure 7). The PCA (for all GP together) explained 81.7% of the variance present using the first four dimensions. Positively correlated variables were CC_75PCT with CC_tm, CH_DET with CH_tm and CH_SNC with CC_SNC. Examples of negatively correlated variables were CC_75PCT and CC_6W (
Figure 6A and
Figure 7E). Variables that show an orthogonal relation in the PCA graphs provide complementary data, for example CH_Hmax and CC_6W, indicating that the maximum height achieved is not correlated with the canopy development in early stages. Based on the contributions of the variables to the two first principal components (
Figure 6A), the most important variables to explain the variability in the data set were CH_tm, CH_te and CC_75PCT for the first principal component and CH_Hmax and CC_AGRmax for the second principal component. The total variance in the PCA is further explained by the CH derived parameters than the CC ones (57.9% versus 42.1%, data not shown). A large overlap between GPs was observed when the plots were plotted in the two first dimensions of the PCA (
Figure 6B) and similar patterns were observed when correlations were calculated per GP (
Figure 7), indicating no clear-cut differentiation among GPs.
SY, R, and R8 values obtained using ground-based methods were plotted on top of the PCA carried out with the phenotypic variables derived from UAV-based data (
Figure 6, red arrows). SY and R8 are positively correlated, both with each other as well as largely with CH_te, CH_tm, and CH_DET. SY and R8 are negatively correlated with CC_SNC and CH_SNC. This is consistent with the results shown in
Figure 7. The low correlation of CC_max with all variables was expected, as all plots reached a high maximum canopy cover, resulting in a small variance. Lower correlations between SY, R1, and R8 with all the other variables were observed for GP4 compared to the other groups (
Figure 7D).
3.4. Linking UAV-Based and Breeder Information
In
Table 4, the results of the four MLR models built for SY and R8 with different sets of variables are presented. Higher adjusted R
2 value and lower RMSE were obtained for R8 compared to SY. The similar adjusted R
2 between the MLR models based on different datasets indicated that SY and R8 could be estimated with similar performance either using the phenotypic parameters (from curve fitting) or the 14 time points (flights), with a trend towards a better prediction based on the raw data, but resulting in models that are less interpretable.
When UAV phenotypic variables were used in the MLR building, the best result for SY was achieved when using three variables: CC_SNC (senescence ratio), CH_te (thermal time when maximum height is reached) and CH_AGRmax (maximum growth rate) with an RMSE of 4.65 g (9% improvement compared to the basic model). In
Figure 8A, the RMSE evolution in response to adding a variable is visualized, whereas
Figure 8B shows the intercept, the coefficients for each GP and the variables included in each model. The red dashed line indicates the best model with three extra variables compared to the basic model.
Figure 8 and
Table 4 demonstrate that the models for SY show stable coefficients for the groups, independently of the number of variables added. This indicates a separate effect explained by the dummy variables. The coefficients for CC_SNC, CH_te and CH_AGRmax in the final model have the same magnitude and have consequently comparable importance. The negative coefficient for CC_SNC indicates that an earlier senescence reduces SY. A positive coefficient for CH_te and CH_AGRmax suggests that a later date to achieve the maximum canopy height and a higher AGRmax are linked with a higher SY In the case of R8, a similar figure is presented and a best model with four phenotypic variables resulted in a RMSE of 63.46 (23% improvement compared to the basic model). The variables included in the model were CC_SNC, CH_Hmax, CH_tm and CC_Cmax. Figures for the other three MLR models are included in the
Supplementary Materials (Figures S4–S6).
In the case of SY, using the 14 time points, five variables were selected as the best model obtaining a RMSE of 4.58 g (11% improvement compared to the basic model). Four of these variables corresponded to CC data. For R8, eight variables were selected including CC and CH data and RMSE was 56.94 (improvement of 31% compared to the basic model).
In a next step, we explored the information obtained in a breeding context to detect any plots displaying interesting traits. For example, by plotting genotypes/plots in terms of relevant complementary trait values (e.g., CH_Hmax, CC_6W and CC_SNC; maximum height reached, fast soil coverage and the rate of senescence, respectively) (
Figure 9A). As apparent in
Figure 9, taller genotypes have a tendency to senesce later. Based on the values obtained, breeders could select a plot with a high CC_6W (good weed suppression), not too tall (low sensitivity to lodging), not too small (strong correlation with seed yield) and a fast senescence. As indicated in
Figure 9B, plot number 30 fulfils these criteria (plots 110, 131, 177, and 392 are also not too tall and display a good weed suppression, but they senesce later).
4. Discussion
4.1. General Assessment of the Methodology Developed
Weekly flights (n = 14) of a UAV equipped with an RGB sensor allowed to phenotype a large field trial during the entire growing season in terms of the dynamics of canopy cover based on the calculation of the vegetation index Excess Green (ExG), and canopy height based on the Structure from Motion (SfM) photogrammetry with high temporal and spatial resolution. These data were used to fit sigmoid curves (Gompertz and Beta function) in an automated fashion using R scripts. Model parameters to objectively describe the different soybean plots/genotypes were extracted. The methodology presented here is suitable for the efficient phenotyping of a large number of genotypes under field conditions and can provide breeders with trait information that may be difficult to obtain using other methods, e.g., maximum absolute growth rate, weed suppressive ability, onset and evolution of senescence or determinacy. Large differences were observed among the four groups considered, and high variability was present within the groups (~genotypes). The full canopy cover was reached starting from day of the year (DOY) 165 (mid-June). A decrease in cover and height was observed starting from DOY 200 (mid/end July) and was related to the onset of senescence.
By fitting a sigmoid growth function such as the Gompertz or the Beta function to multi-temporal data, it is possible to determine reliable parameters with a straightforward biological meaning enabling characterization of between-plot differences in growth and developmental processes [
22]. An extra advantage of curve fitting to multi-temporal data is that possible small errors at one time point are smoothed out using the entire time series. However, model choice is critical because use of the wrong model can introduce bias [
23]. The success rate reached for model fitting was 91.0% for canopy cover and 99.6% for canopy height, hence the models chosen were appropriate for fitting these data. The majority of the cases that did not reach a good fit, originated from CC data mainly from genotypes of GP1, the group first sown. Thus, a bad fit was most likely a combination of having only two data points in the increasing phase of the Gompertz model fit for CC data, the first data point having already a canopy cover of >15%, and the reduced flexibility of the Gompertz function to properly fit the first time points. These first time points are important for the model fit, as these present bigger differences because of rapid growth. This suggests that for UAV based phenotyping using time series, flights should start early enough, at plant emergence, and should last long enough to enable full senescence of all treatments. This sometimes conflicts with traditional experimental methods. In this case, bird netting was used to cover the field trial at the time of emergence to prevent bird damage. This of course interferes with UAV imaging, requiring removal and replacement of large areas of netting at the starting phase of the experiment. Near the end of the experiment, full senescence is not always required in a breeding trial before harvests are performed. Therefore, before starting the experiment, these issues should be discussed carefully between the breeders and the phenotyping researchers. A gap in the dataset can happen too—in this study, no flights were possible between July 25 and August 16. This was solved by the curve fitting, as the values during the two weeks gap were estimated. On July 25 the canopy was fully closed, and on August 16 senescence has been initiated only for some plots. Potentially, this could affect the parameters CC_SNC and CH_SNC, but not all other parameters, as full canopy and maximum height were reached before that gap. Scripting the models in R proved appropriate to automatize the fitting step for large sets of plots. Nevertheless, care should be taken to define appropriate initialization values for the Gompertz model as an optimization was not always achieved with the optimx function in R. In our case different values needed to be tested. Regarding canopy height, some datasets contained close to zero or even negative values for some plots in the first and the second flight. This can be explained by the intrinsic altimetric error and the difficulty of detecting small objects in the photogrammetry process of a flight [
10]. This “noise” cannot be corrected or removed, but is in our opinion acceptable (x-y-z average error is 0.07 m).
4.2. Describing General Growth Patterns in the Different Groups
Using the derived and estimated phenotypic parameters (
Figure 4;
Figure 5), it was possible to detect and quantify differences among the groups. A relevant trait is the canopy cover at six weeks after emergence (CC_6W) as this is related to weed suppression capacity and row closure. In this study, most of the plots of GP3 and GP4 had CC_6W values >90% and closed the row faster than the other two groups. Senescence was also different among groups, with fast senescence for GP4. For CH_AGRmax comparable values were observed for GP3 and GP4 and for GP1 and GP2, but at different thermal times (CH_tm). By combining the results obtained for CH_Hmax and CH_te, we can conclude that plots of GP4 did not grow tall (relatively lower CH_Hmax) but grew faster (lower CH_te in combination with higher CH_AGRmax) than plots in other groups. This confirms our expectations, as GP4 comprised genotypes expected to develop quickly and mature early, according to the information available beforehand. We can thus conclude that the pipeline developed to characterize the seasonal growth and development of soybean is functional and suitable for implementation in real-time breeding applications.
Determinacy describes the extent to which a plant continues to produce extra internodes and consequently extra possibilities to produce pods and seeds after entering the generative phase (R1). Indeterminacy or semi-determinacy can be a favorable trait in soybean for cultivation in Northwest Europe as it represents an ‘insurance policy’ if a period of unfavorable weather conditions occurs, such as low temperature during flowering or drought in the summer months. Based on experience, determinate genotypes perform less well under Northwestern European conditions [
24,
25]. In contrast, a high level of determinacy contributes to a synchronous seed maturation, avoiding variable moisture content in the harvested seed that could cause quality problems. In soybean, determinate cultivars are used in areas with long growing seasons while indeterminate cultivars are more suited to short growing seasons [
26]. Detailed information on this trait is therefore very useful for soybean breeders. Here, we present a method to estimate determinacy in a straightforward way that could be implemented routinely in soybean breeding. Overall, genotypes included in groups 1 and 2 had a higher level of indeterminacy and kept growing during a longer thermal time.
4.3. Estimating SY and Maturity Using UAV-Derived Information
In Northwest Europe, early maturity (quick progress to R8) in combination with high seed yield and a high protein content are important breeding targets. We evaluated if seed yield (SY) and the R8 stage could be predicted by an MLR with the individual time series data points or with the estimated and derived phenotypic parameters as variables. Similar adjusted R2 values were obtained using the data of the different flights and using the derived phenotypic parameters. This demonstrates that the phenotypic parameters captured all variance present. Seed yield is influenced by canopy cover because a longer lasting canopy cover most likely results in more photosynthesis for optimal SY at the end of the growing season. This is supported by the presence of CC_SNC in the first MLR model and because four out of five time points included were CC data for the second model. Canopy height also plays a role in terms of growth rate and the moment when maximum height is reached. An early senescence decreases SY and the later the maximum height is reached, the higher the chances for a higher SY.
Maturity (R8) was determined by more CC time points (five dates, DOY range 164–246) than CH time points (three dates were included). The phenotypic variable CC_SNC has the most influence on R8 followed by CH_tm. Similar results related to maturity prediction classification (93% accuracy) were achieved in Yu et al. 2016 [
12] using RGB and NIR drone data. In the present study we obtained similar results using higher resolution RGB imagery but by using simpler statistical methods (MLR). Soybean agronomic traits such as yield, maturity, and height were predicted in Yuan et al. (2019) [
27] using six flights over four locations by color and texture features. Resulting R
2 values were 0.68, 0.76, and 0.27, respectively, using five regression modeling techniques, i.e., Partial Least Squares Regression, Random Forests, Cubist, Artificial Neural Networks, and Support Vector Regression. In our case, seed yield and R8 estimated by MLR achieved an adjusted R
2 of 0.51 and 0.85, respectively, which is slightly lower for SY but higher for maturity.
4.4. Perspectives for Practical Applications
The UAV-estimated and derived phenotypic data can be used directly in a breeding context, as they are related to relevant traits, e.g., weed suppression capacity, fast canopy establishment, early vigor, growth rates, maximum plant height, risk for lodging, determinacy, etc., for each genotype. For example, maximum plant height contributes negatively to lodging resistance, while it is positively correlated with the number of internodes, which in turn is positively correlated with the number of pods. Therefore, breeders should find a compromise considering different traits. For example, the combination of CC_6W, CH_Hmax, and CC_SNC makes it possible to identify genotypes that combine appropriate values for the different traits.
Besides the use in a breeding context, multi-temporal data can be highly valuable for plant and crop modeling. Time series of different traits have been found to be essential for proper and identifiable parameter estimation [
17]. Large amounts of data can be gathered using UAV with a high temporal resolution.
4.5. Practical Lessons Learned and Future Perspectives
The high spatial resolution achieved from the UAV flights (0.5 cm pixel size for the orthophotos and 1.25 cm for the DEM) revealed the variability present within the plot and yielded a higher number of measurements in comparison to manual data-gathering. High resolution imagery does require a longer processing time, however. To reduce the processing time, a lower spatial resolution could be defined if the aim of the study does not require highly detailed information. Future research could investigate which resolution is sufficient without influencing the predictability of traits.
In this study, we considered 14 flights spread over the entire growing season at regular intervals. This time interval was short in comparison to similar studies, which included an average of only four to 10 dates (e.g., [
13,
16]). During data processing and interpretation, we noticed that a different flight scheme may have been more appropriate. For example, in some cases, as senescence occurred in a short period of time, it could have been appropriate to add to extra flights at the end of the growing season. Similarly, when rapid changes (e.g., fast growth) are expected, extra time points are welcome to ensure a good fit. In this regard, the date of the first flight should also be carefully chosen to capture the entire growing season. In conclusion, planning the flights according to the expected development of the crop may yield more useful data than regular intervals between flights.
5. Conclusions
In this study we collected UAV data of a soybean field trial at an unprecedented temporal resolution. The time series data allowed us to fit growth curves with high accuracy (>90%) and to derive relevant traits. The trait data that became available in this study by combining multi-temporal UAV with curve fitting will allow for a better biological interpretation of the differential response of a large set of soybean genotypes. In addition, we developed a methodology to predict seed yield and to estimate the moment at which each accession reached the R8 stage.
Furthermore, this developed strategy may be applied in the future to the analysis of multi-temporal data obtained using other sensors, e.g., multispectral, hyperspectral and thermal, to evaluate responses to abiotic and biotic stresses. This would enable the development of more resilient plant varieties at a faster pace.