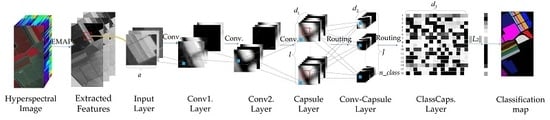
1. Introduction
The task of classification, when it relates to hyperspectral images (HSIs), generally refers to assigning a label to each pixel vector in the scene [
1]. HSI classification is a crucial step for a plethora of applications including urban development [
2,
3,
4], land change monitoring [
5,
6,
7], scene interpretation [
8,
9], resource management [
10,
11], and so on. Due to the fundamental importance of this step in various applications, classification of HSI is one of the hottest topics in the remote sensing community. However, the classification of HSI is still challenging due to several factors such as high dimensionality, a limited number of training samples, and complex imaging situations [
1].
During the last few decades, a huge number of methods have been proposed for HSI classification [
12,
13,
14]. Due to the availability of abundant spectral information in HSIs, lots of spectral classifiers have been proposed for HSI classification including k-nearest-neighbors, maximum likelihood, neural network, logistic regression, and support vector machines (SVMs) [
1,
15,
16].
Hyperspectral sensors provide rich spatial information as well, and the spatial resolution is becoming finer and finer along with the development of sensor technologies. With the help of spatial information, classification performance can be greatly improved [
17]. Among the spectral-spatial classification techniques, the generation of a morphological profile is a widely-used approach, which is usually followed by either an SVM or a random forest classifier to obtain the final classification result [
18,
19,
20,
21]. As the extension of SVM, multiple kernel learning is another main stream of spectral-spatial HSI classification, which has a powerful capability to handle the heterogeneous features obtained by spectral-spatial hyperspectral images [
22].
Due to the complex atmospheric conditions, scattering from neighboring objects, intra-class variability, and varying sunlight intensity, it is very important to extract invariant and robust features from HSIs for accurate classification. Deep learning uses hierarchical models to extract invariant and discriminate features from HSIs in an effective manner and usually leads to accurate classification. During the past few years, many deep learning methods have been proposed for HSI classification. Deep learning includes a broader family of models, including the stacked auto-encoder, the deep belief network, the deep convolutional neural network (CNN), and the deep recurrent neural network. All of the aforementioned deep models have been used for HSI classification [
23,
24].
The stacked auto-encoder was the first deep model to be investigated for HSI feature extraction and classification [
25]. In [
25], two stacked auto-encoders were used to hierarchically extract spectral and spatial features. The extracted invariant and discriminant features led to a better classification performance. Furthermore, recently, the deep belief network was introduced for HSI feature extraction and classification [
26,
27].
Because of the unique and useful model architectures of CNNs (e.g., local connections and shared weights), such networks usually outperform other deep models in terms of classification accuracy. In [
28], a well-designed CNN with five layers was proposed to extract spectral features for accurate classification. In [
29], a CNN-based spectral classifier that elaborately uses pixel-pair information was proposed, and it was shown to obtain good classification performance under the condition of a limited number of training samples.
Most of the existing CNN-based HSI classification methods have been generalized to consider both spectral and spatial information in a single classification framework. The first spectral-spatial classifier based on CNN was introduced in [
30], which was a combination of principal component analysis (PCA), deep CNN, and logistic regression. Due to the fact that the inputs of deep learning models are usually 3D data, it is reasonable to design 3D CNNs for HSI spectral-spatial classification [
31,
32]. Furthermore, CNN can be combined with other powerful techniques to improve the classification performance. In [
33], CNN was combined with sparse representation to refine the learnt features. CNNs can be connected with other spatial feature extraction methods, such as morphological profiles and Gabor filtering, to further improve the classification performance [
34,
35].
The pixel vector of HSIs can be inherently considered to be sequential. Recurrent neural networks have the capability of characterizing sequential data. Therefore, in [
27], a deep recurrent neural network that can analyze hyperspectral pixel vectors as sequential data and then determine information categories via network reasoning was proposed.
Although deep learning models have shown their capabilities for HSI classification, some disadvantages exist which downgrade the performance of such techniques. In general, deep models require a huge number of training samples to reliably train a large number of parameters in their networks. On the other hand, having insufficient training samples is a frequent problem in remotely sensed image classification. In 2017, Sabour et al. proposed a new idea based on capsules, which showed its advantages in coping with a limited number of training samples [
36]. Furthermore, traditional CNNs usually use a pooling layer to obtain invariant features from the input data, but the pooling operation loses the precise positional relationship of features. In hyperspectral remote sensing, abundant spectral information and the positional relationship in a pixel vector are the crucial factors for accurate spectral classification. Therefore, it is important to maintain the precise positional relationship in the feature extraction stage. In addition, when it comes to extracting spectral-spatial features from HSI, it is also important to hold the positional relationship of spectral-spatial features. Moreover, most of the existing deep methods use a scalar value to represent the intensity of a feature. In contrast, capsule networks use vectors to represent features. The usage of vectors enriches the feature representation and is a huge progress and a much more promising method for feature learning than scalar representation [
36,
37]. These properties of the capsule network perfectly align with the goals of this study and the current demands in the hyperspectral community.
Deep learning-based methods, including deep capsule networks, have a powerful feature extraction capability when the number of training samples is sufficient. Unfortunately, the availability of only a limited number of training samples is a common bottleneck in HSI classification. Deep models are often over-trained with a limited number of training samples, which downgrades the classification accuracy on test samples. In order to mitigate the overfitting problem and lessen the feature extraction workload of deep models, the idea of a local connection-based capsule network is proposed in this study. The proposed Conv-Capsule network uses local connections and shared transform matrices to reduce the number of trainable parameters compared to the original capsule, which potentially mitigates the overfitting issue when the number of available training samples is limited.
In the current study, the idea of the capsule network is modified for HSI classification. Two deep capsule classification frameworks, 1D-Capsule and 3D-Capsule, are proposed as spectral and spectral-spatial classifiers, respectively. Furthermore, two modified capsule networks, i.e., 1D-Conv-Capsule and 3D-Conv-Capsule, are proposed to further improve the classification accuracy.
The main contributions of the paper are briefly summarized as follows.
- (1)
A modification of the capsule network named Conv-Capsule is proposed. The Conv-Capsule uses local connections and shared transform matrices in the network, which reduces the number of trainable parameters and mitigates the overfitting issue in classification.
- (2)
Two frameworks, called 1D-Capsule and 3D-Capsule, based on the capsule network are proposed for HSI classification.
- (3)
To further improve the HSI classification performance, two frameworks, called 1D-Conv-Capsule and 3D-Conv-Capsule, are proposed.
- (4)
The proposed methods are tested on three well-known hyperspectral data sets under the condition of having a limited number of training samples.
The rest of the paper is organized as follows.
Section 2 presents the background of the deep learning and capsule network.
Section 3 and
Section 4 are dedicated to the details of the proposed deep capsule network frameworks, including spectral and spectral-spatial architectures for HSI classification. The experimental results are reported in
Section 5. In
Section 6, the conclusions and discussions are presented.
5. Experimental Results
5.1. Data Description
In our study, three widely-used hyperspectral data sets with different environmental settings were used to validate the effectiveness of the proposed methods. They were captured over Salinas Valley in California (Salinas), Kennedy Space Center (KSC) in Florida, and an urban site over the University of Houston campus and the neighboring area (Houston).
The first data set was captured by the 224-band AVIRIS sensor over Salinas Valley, California. After removing the low signal to noise ratio (SNR) bands, the available data set was composed of 204 bands with 512 × 217 pixels. The ground reference map covers 16 classes of interest. The hyperspectral image is of high spatial resolution (3.7-meter pixels).
Figure 5 demonstrates the false-color composite image and the corresponding ground reference map. The number of samples in each class is listed in
Table 3.
The second data set, KSC, was collected by the airborne AVIRIS instrument over the Kennedy Space Center, Florida. The KSC data set has an altitude of approximately 20 km, with a spatial resolution of 18 m. After removing water absorption and low SNR bands, 176 bands with 512 × 614 pixel vectors were used for the analysis. For classification purpose, 13 classes were selected. The classes of the KSC data set and the corresponding false-color composite map are demonstrated in
Figure 6. The number of samples for each class is given in
Table 4.
The third data set is an urban site over the University of Houston campus and neighboring area which was collected by an ITRES-CASI 1500 sensor. The data set is of 2.5-m spatial resolution and consists of 349 × 1905 pixel vectors. The hyperspectral image is composed of 144 spectral bands ranging from 380 to 1050 nm. Fifteen different land-cover classes are provided in the ground reference map, as shown in
Figure 7. The samples are listed in
Table 5.
For all three data sets, we split the labeled samples into three subsets, i.e., training, validation, and test samples. In our experiment, we randomly chose 200 labeled samples as the training set to train the weights and biases of each neuron and transformation matrix between two consecutive capsule layers. The proper architectures of our network were designed based on performance evaluation on 100 validation samples, which were also randomly chosen from labeled samples. The choice of hyper-parameters, like kernel size in the convolution operation and the dimensions of the vector output of each capsule, were also guided by the validation set. After the training was done, all remaining labeled samples served as the test set to evaluate the capability of the network and to obtain the final classification results. Three evaluation criteria were investigated: overall accuracy (OA), average accuracy (AA), and Kappa coefficients (K).
5.2. The Classification Results of the 1D Capsule Network
The 1D capsule network, which is built only based on spectral features, contains two parts. One is a fully connected capsule network that uses normalized spectral vectors as input. The other is the Conv-Capsule network which inputs spectral features extracted by PCA. We call the two methods 1D-Capsule and 1D-Conv-Capsule for short. In the 1D-Conv-Capsule, we first used PCA to reduce the spectral dimensions of the data. Then, we randomly chose 200 and 100 labeled samples as the training and validation data for each data set. The training samples were imported to the 1D capsule network. The number of principal components was chosen based on the classification result for the validation samples. Some other hyper-parameters (e.g., the learning rate, the convolutional kernel size, the in LeakyReLU, etc.) were also determined by the validation set. In our method, the size of the mini-batch was 100 and the number of training epochs was set to 150 for our network. We used a decreasing learning rate which was initialized to 0.01 at the beginning of the training process. The number of the principal components was set to 20, 20, and 30, respectively, for the Salinas, KSC, and Houston data sets. We used in the LeakyReLU function. The parameters , , and in the loss function were set to 0.9, 0.1, and 0.5, respectively.
The main architectures of the 1D-Conv-Capsule network for each data set are shown in
Table 6. Due to the fact that the same number of principal components was chosen as the input, the network for the Salinas and KSC data sets had the same architecture. In
Table 6, (5 × 1 × 8) × 8 in the fourth layer (i.e., transition layer) means that eight channels of convolution with the kernel size of 5 × 1 were used, and each channel output eight feature maps. Thus, the fourth layer output a capsule with eight channels. The fifth layer was a Conv-Capsule layer with eight (i.e., the number of capsule channels output by the fourth layer) channels of capsule input and 16 channels of capsule output. The kernel size was 5 × 1. We used (5 × 1 × 8) × 16 to represent this operation. The last layer was a fully connected capsule layer. All capsules from the fifth layer were connected with
n_class capsules in this layer. The length of the vector output of each capsule in this layer represents the probability of the network’s input belonging to each class. Between consecutive capsule layers in the 1D-Conv-Capsule, three routing iterations were used to determine the coupling coefficients
.
In this set of experiments, our methods were compared with other classical classification methods that are only based on spectral information. These methods included random forest (RF) [
47], multiple layer perceptron (MLP) [
48], linear support vector machine (L-SVM), support vector machine with the radial basis kernel function (RBF-SVM) [
17], recurrent neural network (RNN) [
24], and the convolutional neural network (1D-CNN) [
28]. Furthermore, 1D-PCA-CNN, which has nearly the same architecture as 1D-Conv-Capsule (apart from the capsule layer), was also designed to give a fair comparison. The classification results are shown in
Table 7,
Table 8 and
Table 9.
The experiment setups of the classical classification methods are described as follows. RF was used for classification. A grid search method and four-fold cross-validation were used to define RF’s two key hyper-parameters (i.e., the number of features to consider when looking for the best split (F) and the number of trees (T)). In the experiment, the search ranges of F and T were (5, 10, 15, 20) and (100, 200, 300, 400), respectively. The MLP used in this experiment was a fully connected neural network with one hidden layer. The used MLP contained 64 hidden units. L-SVM is a linear SVM with no kernel function. RBF-SVM uses the radial basis function as the kernel. In L-SVM and RBF-SVM, a grid search method and four-fold cross-validation were also used to define the most appropriate hyper-parameters (i.e.,
for L-SVM and
for RBF-SVM). In this experiment, the search range was exponentially growing sequences of
and
(
,
). A single layer RNN with a gated recurrent unit and the tanh activation function were adopted. The architecture of 1D-CNN was designed as in [
28] and contained an input layer, a convolutional layer, a max-pooling layer, a fully connected layer, and an output layer. The convolutional kernel size and number of kernels were 17 and 20 for all three data sets. The pooling size was 5, 5, and 4 for the Salinas, KSC, and Houston data sets, respectively.
Table 7,
Table 8 and
Table 9 show the classification results obtained when we used the aforementioned experimental settings. All experiments were run ten times with different random training samples. The classification accuracy is given in the form of mean ± standard deviation. The 1D-Conv-Capsule network showed a better performance in terms of accuracy on all three data sets.
For all three data sets, RBF-SVM, which is famous for handling a limited number of training samples, provides competitive classification results. We use the experiments with 200 training samples as an example to discuss the results. For the Salinas data set, 1D-Conv-Capsule exhibited the best OA, AA, and K, with improvements of 2.05%, 3.01%, and 0.023 over RBF-SVM, respectively. Our approach outperformed 1D-PCA-CNN by 1.6%, 1.62%, and 0.0179 in terms of OA, AA, and K, respectively. For the KSC data set, as can be seen, the OA of 1D-Conv-Capsule was 88.22%, which is an increase of 1.58% and 2.2% compared with RBF-SVM and 1D-PCA-CNN, respectively. For the Houston data set, 1D-Conv-Capsule improved the OA, AA, and K of 1D-PCA-CNN by 3.21%, 2.84%, and 0.0346, respectively. The results show that the 1D-Conv-Capsule method demonstrated the best performance in terms of OA, AA, and K for all three data sets. In addition, all experiments with 100 and 300 training samples were also implemented to demonstrate the effectiveness of the proposed methods. From the results reported in
Table 7,
Table 8 and
Table 9, it can be seen that 1D-Conv-Capsule outperformed the other classical classification methods, especially when the number of training samples was extremely limited (i.e., 100 training samples).
Furthermore, the 1D-Conv-Capsule with a different number of principal components as input was conducted.
Figure 8 shows the classification results of the 1D-Conv-Capsule on three data sets by using 200 training samples. Due to the fact that we injected only spectral information into the 1D-Conv-Capsule, relatively more principal components were used to make sure that sufficient spectral information was preserved, and, at the same time, this maintained low computational complexity. From
Figure 8, it can be seen that if the number of selected components is too small or too big, the classification results tend to be poor under both circumstances. On one hand, the spectral information is not sufficiently preserved and the network cannot efficiently extract the spectral feature when the number of principal components is low. On the other hand, the networks are over-trained when the number of principal components is high. The situation becomes worse if the number of training samples is limited. The best classification performance was achieved when the number of the principal components was set to 20, 20, and 30 for the Salinas, KSC, and Houston data sets, respectively.
5.3. The Analysis of Learnt Features of the 1D Capsule
From the aforementioned description about the capsule, it can be understood that the output of the capsule is a vector representation of the type of entity. In order to demonstrate the real advantage of the capsule network on remote sensing data, we performed another experiment based on the 1D-Capsule network followed by a reconstruction network (1D-Capsule-Recon). The architecture of the reconstruction network is shown in
Figure 9. According to the label of the input pixel, the representative vector of the corresponding capsule in the ClassCaps layer was imported to the reconstruction network (e.g., if the input pixel belonged to the
-th class, the vector output of the
-th capsule in the ClassCaps layer was used as input to the reconstruction network). The reconstruction network contained three fully connected (FC) layers. The first two FC layers had 128 and 256 hidden units with the ReLU activation function. The last FC layer with Sigmoid activation function output the reconstructed spectra (i.e., a combination of normalized spectral reflectance of different bands) corresponding to the input of the 1D-Capsule-Recon. The reconstruction loss, i.e., the Euclidean distance between the input and the reconstructed spectra, was added to the margin loss that described in
Section 3:
where
is the margin loss and
is the reconstruction loss.
is the weight coefficient that is used to avoid
dominating
during the training procedure. In the experiment,
was set to 0.1.
was used as the loss function for the 1D-Capsule-Recon.
To visualize the vector representation of the capsule, we made use of the reconstruction network. After the training procedure of the 1D-Capsule-Recon was done, we randomly chose some samples from different classes and computed the representation vector of their corresponding capsules in the ClassCaps layer. We made perturbations in different dimensions of the vector and fed them to the reconstruction network.
Figure 10 shows the reconstructed results of three class samples from the Salinas data set. Two dimensions of the representation vector were tuned. In
Figure 10, the original is the input spectra to the 1D-Capsule-Recon. The notation of [
] in
Figure 10 means that we tuned the
-th dimension of the representation vector
with perturbation
. The perturbed
was used to reconstruct the spectra. From the results shown in
Figure 10, the representation vector (i.e.,
) can well reconstruct the spectra, which means that the representation vector contains the information in the spectra with low dimensionality.
Furthermore, as shown in
Figure 10,
can influence the reconstruction of some special bands, which means that
has a close relationship with the special bands.
is a vector that contains several
, and
is a robust and condensed representation of spectra.
5.4. The Classification Results of the 3D Capsule Network
In the 3D capsule network, the network extracts both spectral and spatial features effectively, which could lead to a better performance in terms of classification accuracy than the one obtained by the 1D capsule network. As mentioned above, we proposed two 3D frameworks, i.e., the 3D-Capsule and the 3D-Conv-Capsule. Similar to a 1D framework, the 3D-Capsule is an original fully connected capsule network, while the 3D-Conv-Capsule is the convolutional capsule network. Additionally, the 3D-Capsule directly uses the original hyperspectral data as input, while the 3D-Conv-Capsule utilizes EMAP to extract features of hyperspectral data. In the 3D-Conv-Capsule, three principal components were used and parameters in EMAP were set as in [
21]. Through the EMAP analysis, the number of spectral dimensions became 108 for all three data sets. In this set of experiments, the numbers of training and validation samples were the same as for the 1D Capsule network. The mini-batch size was also 100. The training epoch was set to 100 with a learning rate of 0.001. The parameter in loss function was the same as for the 1D capsule network. The details on the architecture of the 3D-Conv-Capsule network are shown in
Table 10. The definitions of the parameters in
Table 10 can be found in the description for the 1D-Conv-Capsule network. Batch normalization was also used to improve the performance of the network.
The SVM-based and CNN-based methods were included in the experiments to give a comprehensive comparison. The classification results are shown in
Table 11,
Table 12 and
Table 13. For the three data sets, we used 27 × 27 neighbors of each pixel as input 3D images in these methods.
Due to the high performance in terms of classification accuracy of SVM, some SVM-based HSIs classifiers were adopted for comparison. The extended morphological profile with SVM (EMP-SVM) is a widely used spectral-spatial classifier [
19]. In the EMP-SVM method, the morphological opening and closing operations were used to extract spatial information on the first three components of HSIs, which were computed by PCA. In the experiments, the shape structuring element (SE) was set as a disk, and the radius of disk increased from two to eight with an interval of two. Therefore, 27 spatial features were generated. The learned features were fed to an RBF-SVM to obtain the final classification results. EMAP is a generalization of the EMP and can extract more informative spatial information. EMAP was also combined with the random forest classifier (EMAP-RF) [
20]. In order to have a fair comparison, the parameters in EMAP were kept the same as for the 3D-Conv-Capsule. In RBF-SVM, the optimal parameters
and
were also obtained by grid-search and four-fold cross-validation methods. Furthermore, CNN was also used for comparison. We conducted 3D-CNN, EMP-CNN and 3D-EMAP-CNN. Their CNN architectures were the same as in [
31]. To give a comprehensive comparison, a spectral–spatial residual network recently proposed in [
49] was adopted for comparison.
Table 11,
Table 12 and
Table 13 give the classification results of the proposed methods and contrast methods on the three data sets. We also used the classification results with 200 training samples as an example. For the Salinas data set, the 3D-Conv-Capsule exhibited the highest OA, AA, and K, with the improvements of 3.64%, 3.35%, and 0.0409 over 3D-EMAP-CNN, respectively. On the other hand, our 3D-Capsule approach also performed better than 3D-EMAP-CNN in terms of OA, AA, and K. For the KSC data set, 3D-Conv-Capsule improved the OA, AA, and K of EMP-CNN by 2.19%, 3.36%, and 0.0244, respectively. Our 3D-Capsule method also showed higher classification accuracy than EMP-CNN with improvements of 0.52%, 1.12%, and 0.0057 in terms of OA, AA, and K. For the Houston data set, we obtained similar results. Experiments with 100 and 300 training samples were investigated as well. The detailed classification results are shown in
Table 11,
Table 12 and
Table 13. Compared with other state-of-the-art methods, the 3D-Conv-Capsule demonstrated the best performance under different training samples.
In the experiment using the 3D-Conv-Capsule, we also explored how a different number of principal components that are used in EMAP analysis may affect the classification results. Due to the spatial information being considered and the EMAP analysis significantly increasing the data volume, we used relatively fewer principal components here compared with the 1D-Conv-Capsule.
Figure 11 shows the classification result for the 3D-Conv-Capsule. The 3D-Conv-Capsule with different numbers of principal components outperformed the other contrast experiments. Unlike 1D-Conv-Capsule, the preservation of more principal components leads to a vast data volume which brings a higher requirement for hardware and longer training time in 3D-Conv-Capsule. Though the classification accuracy may be higher with relatively more components, we only used three principal components in consideration of computational cost in the 3D-Conv-Capsule.
5.5. Parameter Analysis
In the 3D-Conv-Capsule, convolutional layers were used as feature extractors, and they converted the original input into a capsule’s input. Thus, the number of convolutional layers and the convolutional kernel size used in 3D-Conv-Capsule influences the classification performance of the model. Furthermore, due to the fact that the input of a 3D-Conv-Capsule is the a × a neighbors around the pixel, the size of neighborhoods is also an important factor. These factors are analyzed below.
When we explored the influence of a parameter on the classification result, the other parameters were fixed. The neighborhood size and convolution kernel size were set to 27 and 3 when we analyzed the number of convolutional layers. For the analysis of the convolution kernel size, 27 × 27 neighborhoods and two convolutional layers were used in the 3D-Conv-Capsule. Similarly, the number of convolutional layers and the convolution kernel size were set to 2 and 3 for analysis of the size of the neighborhood. All the experiments for this analysis were conducted with 200 training samples.
Table 14,
Table 15 and
Table 16 shows the detailed classification results. As reported in
Table 14, the use of two convolutional layers gave better classification results. Furthermore, one convolutional layer could not extract features efficiently while three layers made the model prone to overfitting.
Table 15 shows the classification results with different convolution kernel sizes. The 3D-Conv-Capsule performed better when the kernel size was 3. For the neighborhood size, the 3D-Conv-Capsule obtained good classification accuracies on the Salinas and KSC data sets when the neighborhood size was relatively large, but the result for the Houston data set was the other way around.
5.6. Visualization of Learnt Features from the Capsule Network
Unlike traditional neural networks which use a sequence of scalar value to represent the probability of the input belonging to different classes, capsule networks output n_class (i.e., the number of classes) capsules that represent different classes of entity. The length of the vector output of each capsule (i.e., the Euclidean norm of the vector) represents the probability that a corresponding entity exists. In HSI classification tasks, the length of different capsules’ output vectors can be interpreted as the probability that the input belongs to different classes.
We randomly choose several samples from the test data set and imported them into the trained 3D-Conv-Capsule network. The length of the vector output of each capsule in the ClassCaps layer is computed and visualized in
Figure 12. From the results shown in
Figure 12, it is possible to observe that the capsule corresponding to the true class output the longest vector. Due to the similarity between the Graminoid marsh and Spartina marsh, the experimental results of three samples from the Graminoid marsh class show that the length of the vector corresponding to the similar class was longer than those of the other classes.
5.7. Time Consumption
All experiments in this paper were conducted on a Dell laptop equipped with an Intel Core i5-7300H processor with 2.5 GHz, 8 GB of DDR4 RAM, and an NVIDIA GeForce GTX 1050Ti graphical processing unit (GPU). The software environment used Windows 10 as an operating system, CUDA 9.0 and cuDNN 7.1, Keras framework using TensorFlow as a backend, and Python 3.6 as the programing language. The training and test times of different models are reported in
Table 17 and
Table 18. The traditional RF and SVM classifiers demonstrated superior computational efficiency. As for deep learning models, the model was able to be trained within a few minutes due to the limited number of training samples and the GPU’s strong computing acceleration power. The 3D-Conv-Capsule required nearly the same training time as 3D-CNN and less time than SSRN. In the experiments, it was found that capsule network-based method converged “faster” than the CNN-based method (e.g., 100 epochs for 3D-Conv-Capsule and 500 epochs for 3D-EMAP-CNN). In future work, the use of more specific computing acceleration for the capsule network could further boost the computational efficiency of the capsule-based method.
5.8. Classification Maps
Lastly, we evaluated the classification accuracies from a visual perspective. The trained models, including 1D-CNN, 1D-Conv-Capsule, EMAP-SVM, 3D-CNN and 3D-Conv-Capsule, were selected to classify the whole images. All parameters in these models were optimized.
Figure 13,
Figure 14 and
Figure 15 show the classification maps obtained by different models using the three data sets. From
Figure 13,
Figure 14 and
Figure 15, we can figure out how the different classification methods affect the classification results. Although the 1D-Conv-Capsule demonstrated a higher accuracy than 1D-CNN, the 1D-CNN and 1D-Conv-Capsule models, which only utilize spectral features, depicted more errors compared with spectral-spatial-based methods for the three data sets. Spectral-based models always result in noisy scatter points in the classification map (see
Figure 13b,c,
Figure 14b,c and
Figure 15b,c). Spectral-spatial methods overcome this shortcoming. Obviously, 3D-CNN and 3D-Conv-Capsule, which directly use the neighbor information as the model input, resulted in smoother classification maps. By comparing the true ground reference with the classification maps, the 3D-Conv-Capsule obtained more precise classification results, which demonstrates that the capsule network is an effective method for HSI classification.
6. Conclusions
In this paper, an improved capsule network called the convolutional capsule (Conv-Capsule) was proposed. On the basis of Conv-Capsule, new deep models called 1D-Conv-Capsule and 3D-Conv-Capsule were investigated for HSI classification. Furthermore, 1D-Conv-Capsule and the 3D-Conv-Capsule were combined with PCA and EMAP, respectively, to further improve the classification performance.
The proposed models, 1D-Conv-Capsule and 3D-Conv-Capsule, can effectively extract spectral and spectral-spatial features from HSI data. They were tested on three widely-used hyperspectral data sets under the condition of having a limited number of training samples. The experimental results showed the superiority over the classical SVM-based and CNN-based methods in terms of classification accuracy.
The proposed methods explored the convolutional capsule network for HSI classification, representing a new methodology for better modeling and processing of HSI. Compared with a fully connected capsule layer, the convolutional capsule layer dramatically reduces the trainable parameters, which is critical in order to avoid over-training. In our future work, based on the convolutional capsule, deep capsule architecture like SSRN in CNN will be conducted to fully investigate the potential of capsule networks.