A 3D Point Cloud Filtering Method for Leaves Based on Manifold Distance and Normal Estimation
Abstract
:1. Introduction
1.1. Background
1.2. Objectives
2. Materials and Methods
2.1. Data Collection by TLS
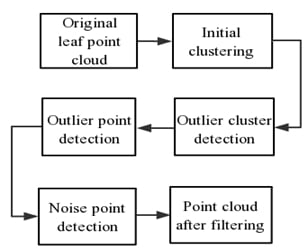
2.2. Framework of the Proposed Method
2.3. Adapted Truncation Method
2.4. Outlier Detection and Filtering in Manifold Space
2.4.1. Measuring Manifold Distance
2.4.2. Filtering Outlier Clusters
| Input: Point cloud |
|
| Output: New clusters without outlier clusters: , |
2.4.3. Filtering Outlier Points
| Input: , , , |
|
| Output: Clusters without outlier points: , |
2.5. Noise Detection and Filtering Based on the Normal Feature of Local Surfaces
2.5.1. Local Surface Fitting and Normal Estimation
2.5.2. Filtering Noise Points
| Input: , , , |
|
| Output: Cluster without outlier and noise points, , |
3. Experiments and Results Analysis
3.1. Effects of the Threshold Value on Recognition Rates
3.2. Filtering Performance Evaluation Based on 3D Reconstruction
3.3. Leaf Area Measurement
4. Discussion
4.1. Comparison with Existing Methods
4.2. Recommendations
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Giuliani, R.; Magnanini, E.; Nerozzi, F.; Muzzi, E.; Sinoquet, H. Canopy probabilistic reconstruction inferred from monte carlo point-intercept leaf sampling. Agric. For. Meteorol. 2005, 128, 17–32. [Google Scholar] [CrossRef]
- Chaivivatrakul, S.; Tang, L.; Dailey, M.N.; Nakarmi, A.D. Automatic morphological trait characterization for corn plants via 3D holographic reconstruction. Comput. Electron. Agric. 2014, 109, 109–123. [Google Scholar] [CrossRef]
- Zhu, X.; Zhu, M.; Ren, H. Method of plant leaf recognition based on improved deep convolutional neural network. Cogn. Syst. Res. 2018, 52, 223–233. [Google Scholar] [CrossRef]
- Richter, R.; Reu, B.; Wirth, C.; Doktor, D.; Vohland, M. The use of airborne hyperspectral data for tree species classification in a species-rich Central European forest area. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 464–474. [Google Scholar] [CrossRef]
- Oveland, I.; Hauglin, M.; Gobakken, T.; Næsset, E.; Maalen-Johansen, I. Automatic estimation of tree position and stem diameter using a moving terrestrial laser scanner. Remote Sens. 2017, 9, 350. [Google Scholar] [CrossRef]
- Liu, C.; Xing, Y.; Duanmu, J.; Tian, X. Evaluating different methods for estimating diameter at breast height from terrestrial laser scanning. Remote Sens. 2018, 10, 513. [Google Scholar] [CrossRef]
- Saremi, H.; Kumar, L.; Stone, C.; Melville, G.; Turner, R. Sub-compartment variation in tree height, stem diameter and stocking in a pinus radiata D. don plantation examined using airborne LiDAR data. Remote Sens. 2014, 6, 7592–7609. [Google Scholar] [CrossRef]
- Yun, T. A novel approach for retrieving tree leaf area from ground-based LiDAR. Remote Sens. 2016, 8, 942. [Google Scholar] [CrossRef]
- Bailey, B.N.; Ochoa, M.H. Semi-direct tree reconstruction using terrestrial LiDAR point cloud data. Remote Sens. Environ. 2018, 208, 133–144. [Google Scholar] [CrossRef]
- Berger, M.; Tagliasacchi, A.; Seversky, L.; Alliez, P.; Levine, J.; Sharf, A.; Silva, C. State of the art in surface reconstruction from point clouds. In Eurographics 2014—State of the Art Reports; EUROGRAPHICS Star Reports; The Eurographics Association: Geneva, Switzerland, 2014; Volume 1, pp. 161–185. [Google Scholar]
- Buades, A.; Coll, B.; Morel, J.M. A review of image denoising algorithms, with a new one. Siam J. Multiscale Model. Simul. 2005, 4, 490–530. [Google Scholar] [CrossRef]
- Thakran, Y.; Toshniwal, D. Unsupervised outlier detection in streaming data using weighted clustering. In Proceedings of the International Conference on Intelligent Systems Design & Applications, Kochi, India, 27–29 November 2012. [Google Scholar]
- Schall, O.; Belyaev, A.; Seidel, H.P. Robust filtering of noisy scattered point data. In Proceedings of the Eurographics/IEEE Vgtc Conference on Point-Based Graphics, Stony Brook, NY, USA, 21–22 June 2005; pp. 71–77. [Google Scholar]
- Zaman, F.; Wong, Y.P.; Ng, B.Y. Density-based denoising of point cloud. In Proceedings of the 9th International Conference on Robotic, Vision, Signal, Processing and Power Applications; Springer: Singapore, 2017. [Google Scholar]
- Huang, D.; Du, S.; Li, G.; Zhao, C.; Deng, Y. Detection and monitoring of defects on three-dimensional curved surfaces based on high-density point cloud data. Precis. Eng. 2018, 53, 79–95. [Google Scholar] [CrossRef]
- Narváez, E.A.L.; Narváez, N.E.L. Point cloud denoising using robust principal component analysis. In GRAPP 2006, Proceedings of the First International Conference on Computer Graphics Theory and Applications, Setúbal, Portugal, 25–28 February 2006; DBLP: Trier, Germany, 2016; pp. 51–58. [Google Scholar]
- Zhu, H.; Zhang, J.; Wang, Z. Arterial spin labeling perfusion MRI signal denoising using robust principal component analysis. J. Neurosci. Methods 2017, 295, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, S.M.; Lotfi, A.; Langensiepen, C. User activities outlier detection system using principal component analysis and fuzzy rule-based system. In Proceedings of the International Conference on Pervasive Technologies Related to Assistive Environments, Crete, Greece, 6–9 June 2012; pp. 1–8. [Google Scholar]
- Liao, B.; Xiao, C.; Jin, L.; Fu, H. Efficient feature-preserving local projection operator for geometry reconstruction. CAD Comput. Aided Des. 2013, 45, 861–874. [Google Scholar] [CrossRef] [Green Version]
- Ye, M.; Yang, R.; Pollefeys, M. Accurate 3D pose estimation from a single depth image. In Proceedings of the International Conference on Computer Vision, Barcelona, Spain, 6–13 November 2011; IEEE Computer Society: Washington, DC, USA, 2011; pp. 731–738. [Google Scholar]
- Liu, C.; Li, J.; Zhang, S.; Ding, L. A point clouds filtering algorithm based on grid partition and moving least squares. Procedia Eng. 2012, 28, 476–482. [Google Scholar]
- Fleishman, S.; Cohen-Or, D.; Silva, C.T. Abstract Robust moving least-squares fitting with sharp features. ACM Trans. Graph. (TOG) 2005, 24, 544–552. [Google Scholar] [CrossRef]
- Morigi, S.; Rucci, M.; Sgallari, F. Nonlocal surface fairing. In Scale Space and Variational Methods in Computer Vision; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Maximo, A.; Patro, R.; Varshney, A.; Farias, R. A robust and rotationally invariant local surface descriptor with applications to non-local mesh processing. Graph. Models 2011, 73, 231–242. [Google Scholar] [CrossRef]
- Xing, X.X.; Zhou, Y.L.; Adelstein, J.S.; Zuo, X.N. PDE-based spatial smoothing: A practical demonstration of impacts on MRI brain extraction, tissue segmentation and registration. Magn. Reson. Imaging 2011, 29, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Lai, R.; Liang, J.; Zhao, H.K. A local mesh method for solving PDEs on point clouds. Inverse Probl. Imaging 2014, 7, 737–755. [Google Scholar] [CrossRef]
- Buades, A.; Coll, B.; Morel, J.M. A non-local algorithm for image denoising. In Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05), San Diego, CA, USA, 20–25 June 2005. [Google Scholar]
- Mahmoudi, M.; Sapiro, G. Fast image and video denoising via nonlocal means of similar neighborhoods. IEEE Signal Process. Lett. 2005, 12, 839–842. [Google Scholar] [CrossRef] [Green Version]
- Dabov, K.; Foi, A.; Katkovnik, V.; Egiazarian, K. Image denoising by sparse 3-D transform-domain collaborative filtering. IEEE Trans. Image Process. 2007, 16, 2080–2095. [Google Scholar] [CrossRef]
- Prasath, V.B.S. Image denoising by anisotropic diffusion with inter-scale information fusion. Pattern Recognit. Image Anal. 2017, 27, 748–753. [Google Scholar] [CrossRef]
- Lozes, F.; Elmoataz, A.; Lezoray, O. Partial difference operators on weighted graphs for image processing on surfaces and point clouds. IEEE Trans. Image Process. 2014, 23, 3896–3909. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Cao, L. Tree-Species Classification in Subtropical Forests Using Airborne Hyperspectral and LiDAR Data. Remote Sens. 2017, 9, 1180. [Google Scholar] [CrossRef]
- Maimon, O.; Rokach, L. Data Mining and Knowledge Discovery Handbook; Springer: Boston, MA, USA, 2005. [Google Scholar]
- Hawkins, D.M. Identification of Outliers; Chapman and Hall: London, UK, 1980; pp. 321–328. [Google Scholar]
- Barnett, V.; Lewis, T.; Abeles, F. Outliers in Statistical Data; Wiley & Sons: Hoboken, NJ, USA, 1994; pp. 117–118. [Google Scholar]
- Acuna, E.; Rodriguez, C. A Meta Analysis Study of Outlier Detection Methods in Classification; Department of Ijser Mathematics University of Puerto: San Juan, Puerto Rico, 2004. [Google Scholar]
- Narvaez, P.; Siu, K.Y.; Tzeng, H.Y. New dynamic algorithms for shortest path tree computation. IEEE/ACM Transa. Netw. 2000, 8, 734–746. [Google Scholar] [CrossRef] [Green Version]
- Wei, D. An optimized floyd algorithm for the shortest path problem. J. Netw. 2010, 5, 1496–1504. [Google Scholar] [CrossRef]
- Aini, A.; Salehipour, A. Speeding up the Floyd–Warshall algorithm for the cycled shortest path problem. Appl. Math. Lett. 2012, 25, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Nurunnabi, A.; West, G.; Belton, D. Outlier detection and robust normal-curvature estimation in mobile laser scanning 3D point cloud data. Pattern Recognit. 2015, 48, 1404–1419. [Google Scholar] [CrossRef]
- Hubert, M.; Rousseeuw, P.J.; Branden, K.V. ROBPCA: A new approach to robust principal component analysis. Technometrics 2005, 47, 64–79. [Google Scholar] [CrossRef]
- Rousseeuw, P.J.; Driessen, K.V. A fast algorithm for the minimum covariance determinant estimator. Technometrics 1999, 41, 212–223. [Google Scholar] [CrossRef]
- Nurunnabi, A.; Belton, D.; West, G. Diagnostic-robust statistical analysis for local surface fitting in 3d point cloud data. In Proceedings of the Xxii Congress of International Society for Photogrammetry and Remote Sensing, Melbourne, Australia, 5 August–1 September 2012; pp. 269–274. [Google Scholar]
- Ghasemi, H.; Brighenti, R.; Zhuang, X.Y.; Muthu, J.; Rabczuk, T. Optimization of fiber distribution in fiber reinforced composite by using NURBS functions. Comput. Mater. Sci. 2014, 83, 463–473. [Google Scholar] [CrossRef]
- Hu, C.H.; Li, P.P.; Pan, Z. Phenotyping of poplar seedling leaves based on a 3D visualization method. Int. J. Agric. Biol. Eng. 2018, 11, 145–151. [Google Scholar]
- Sridhar, R.; Rajeev, R.; Kyuseok, S. Efficient algorithms for mining outliers from large data sets. ACM SIGMOD Rec. 2000, 29, 427–438. [Google Scholar] [Green Version]
- Breunig, M.M.; Kriegel, H.P.; Ng, R.T. LOF: Identifying density-based local outliers. In Proceedings of the ACM SIGMOD International Conference on Management of Data, Dallas, TX, USA, 15–18 May 2000; ACM: New York, NY, USA, 2000; pp. 93–104. [Google Scholar]
- Schall, O.; Belyaev, A.; Seidel, H. Adaptive feature-preserving non-local denoising of static and time-varying range data. Comput. Aided Des. 2008, 40, 701–707. [Google Scholar] [CrossRef]
- Han, X.F.; Jin, J.S.; Wang, M.J.; Jiang, W.; Gao, L.; Xiao, L. A review of algorithms for filtering the 3D point cloud. Signal Process. Image Commun. 2017, 57, 103–112. [Google Scholar] [CrossRef]









| Minimum | Maximum | Mean | Standard Deviation | |
|---|---|---|---|---|
| Tree height (m) | 0.71 | 10.8 | 6.2 | 3.6 |
| Tree DBH (cm) | 1.2 | 20.6 | 12.7 | 5.1 |
| Leaf point Number | 189 | 752 | 423 | 112 |
| Leaf length (cm) | 3.4 | 12.4 | 7.1 | 1.9 |
| Leaf width (cm) | 2.7 | 10.5 | 6.2 | 1.8 |
| Filtering Using The Proposed Method | Filtering Using Classical PCA | |||||
|---|---|---|---|---|---|---|
| Leaf Size | SF | MF | LF | SF | MF | LF |
| MAE (cm2) | 0.92 | 1.05 | 3.39 | 2.76 | 5.61 | 6.83 |
| MAE% | 10.63 | 4.83 | 3.8 | 33.68 | 27.05 | 11.79 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, C.; Pan, Z.; Li, P. A 3D Point Cloud Filtering Method for Leaves Based on Manifold Distance and Normal Estimation. Remote Sens. 2019, 11, 198. https://doi.org/10.3390/rs11020198
Hu C, Pan Z, Li P. A 3D Point Cloud Filtering Method for Leaves Based on Manifold Distance and Normal Estimation. Remote Sensing. 2019; 11(2):198. https://doi.org/10.3390/rs11020198
Chicago/Turabian StyleHu, Chunhua, Zhou Pan, and Pingping Li. 2019. "A 3D Point Cloud Filtering Method for Leaves Based on Manifold Distance and Normal Estimation" Remote Sensing 11, no. 2: 198. https://doi.org/10.3390/rs11020198