Comprehensive Evaluation of Lipid Nanoparticles and Polyplex Nanomicelles for Muscle-Targeted mRNA Delivery
Abstract
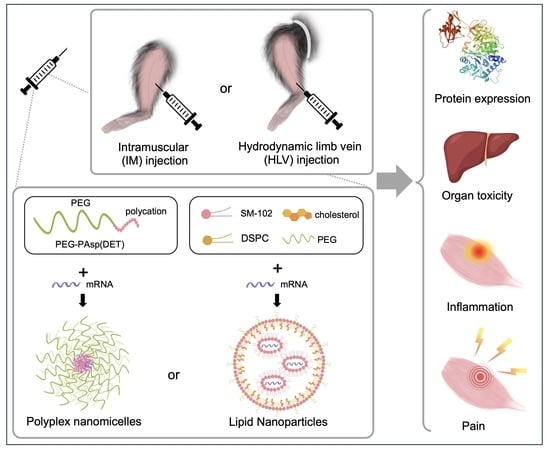
:1. Introduction
2. Materials and Methods
2.1. Preparation of mRNA-Loaded LNP and Polyplex Nanomicelle
2.2. Animal Experiments
2.3. In Vivo Imaging
2.4. Tissue Luciferase Assay
2.5. Blood Biochemistry
2.6. Histology and Image Analysis
2.7. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.8. Static Weight-Bearing Testing
2.9. Statistical Analysis
3. Results
3.1. Evaluation of Luciferase Expression by In Vivo Imaging
3.2. Evaluation of Liver and Kidney Toxicity
3.3. Evaluation of Tissue Damage after Administration
3.4. Evaluation of Inflammatory Response
3.5. Evaluation of the Pain Level
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef]
- Sahin, U.; Karikó, K.; Türeci, Ö. mRNA-based therapeutics—Developing a new class of drugs. Nat. Rev. Drug Discov. 2014, 13, 759–780. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Kong, N.; Zhang, X.; Cao, Y.; Langer, R.; Tao, W. The landscape of mRNA nanomedicine. Nat. Med. 2022, 28, 2273–2287. [Google Scholar] [CrossRef] [PubMed]
- Nitika; Wei, J.; Hui, A.-M. The Delivery of mRNA Vaccines for Therapeutics. Life 2022, 12, 1254. [Google Scholar] [CrossRef]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. [Google Scholar] [CrossRef] [PubMed]
- Feldman, R.A.; Fuhr, R.; Smolenov, I.; Ribeiro, A.; Panther, L.; Watson, M.; Senn, J.J.; Smith, M.; Almarsson, Ö.; Pujar, H.S.; et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine 2019, 37, 3326–3334. [Google Scholar] [CrossRef]
- Aldrich, C.; Leroux–Roels, I.; Huang, K.B.; Bica, M.A.; Loeliger, E.; Schoenborn-Kellenberger, O.; Walz, L.; Leroux-Roels, G.; von Sonnenburg, F.; Oostvogels, L. Proof-of-concept of a low-dose unmodified mRNA-based rabies vaccine formulated with lipid nanoparticles in human volunteers: A phase 1 trial. Vaccine 2021, 39, 1310–1318. [Google Scholar] [CrossRef]
- Burris, H.A.; Patel, M.R.; Cho, D.C.; Clarke, J.M.; Gutierrez, M.; Zaks, T.Z.; Frederick, J.; Hopson, K.; Mody, K.; Binanti-Berube, A.; et al. A phase I multicenter study to assess the safety, tolerability, and immunogenicity of mRNA-4157 alone in patients with resected solid tumors and in combination with pembrolizumab in patients with unresectable solid tumors. J. Clin. Oncol. 2019, 37, 2523. [Google Scholar] [CrossRef]
- Ndeupen, S.; Qin, Z.; Jacobsen, S.; Bouteau, A.; Estanbouli, H.; Igyártó, B.Z. The mRNA-LNP platform’s lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. iScience 2021, 24, 103479. [Google Scholar] [CrossRef]
- Kedmi, R.; Ben-Arie, N.; Peer, D. The systemic toxicity of positively charged lipid nanoparticles and the role of Toll-like receptor 4 in immune activation. Biomaterials 2010, 31, 6867–6875. [Google Scholar] [CrossRef]
- Sedic, M.; Senn, J.J.; Lynn, A.; Laska, M.; Smith, M.; Platz, S.J.; Bolen, J.; Hoge, S.; Bulychev, A.; Jacquinet, E.; et al. Safety Evaluation of Lipid Nanoparticle–Formulated Modified mRNA in the Sprague-Dawley Rat and Cynomolgus Monkey. Vet. Pathol. 2018, 55, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Aini, H.; Itaka, K.; Fujisawa, A.; Uchida, H.; Uchida, S.; Fukushima, S.; Kataoka, K.; Saito, T.; Chung, U.-I.; Ohba, S. Messenger RNA delivery of a cartilage-anabolic transcription factor as a disease-modifying strategy for osteoarthritis treatment. Sci. Rep. 2016, 6, 18743. [Google Scholar] [CrossRef]
- Deng, J.; Fukushima, Y.; Nozaki, K.; Nakanishi, H.; Yada, E.; Terai, Y.; Fueki, K.; Itaka, K. Anti-Inflammatory Therapy for Temporomandibular Joint Osteoarthritis Using mRNA Medicine Encoding Interleukin-1 Receptor Antagonist. Pharmaceutics 2022, 14, 1785. [Google Scholar] [CrossRef]
- Crowley, S.T.; Fukushima, Y.; Uchida, S.; Kataoka, K.; Itaka, K. Enhancement of Motor Function Recovery after Spinal Cord Injury in Mice by Delivery of Brain-Derived Neurotrophic Factor mRNA. Mol. Ther. Nucleic Acids 2019, 17, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Kuniishi, H.; Sakai, K.; Fukushima, Y.; Du, X.; Yamashiro, K.; Hori, K.; Imamura, M.; Hoshino, M.; Yamada, M.; et al. Brain Dp140 alters glutamatergic transmission and social behaviour in the mdx52 mouse model of Duchenne muscular dystrophy. Prog. Neurobiol. 2022, 216, 102288. [Google Scholar] [CrossRef]
- Le Guen, Y.T.; Le Gall, T.; Midoux, P.; Guégan, P.; Braun, S.; Montier, T. Gene transfer to skeletal muscle using hydrodynamic limb vein injection: Current applications, hurdles and possible optimizations. J. Gene Med. 2020, 22, e3150. [Google Scholar] [CrossRef] [PubMed]
- Itaka, K.; Osada, K.; Morii, K.; Kim, P.; Yun, S.-H.; Kataoka, K. Polyplex nanomicelle promotes hydrodynamic gene introduction to skeletal muscle. J. Control. Release 2010, 143, 112–119. [Google Scholar] [CrossRef]
- Kenjo, E.; Hozumi, H.; Makita, Y.; Iwabuchi, K.A.; Fujimoto, N.; Matsumoto, S.; Kimura, M.; Amano, Y.; Ifuku, M.; Naoe, Y.; et al. Low immunogenicity of LNP allows repeated administrations of CRISPR-Cas9 mRNA into skeletal muscle in mice. Nat. Commun. 2021, 12, 7101. [Google Scholar] [CrossRef]
- Sebestyén, M.G.; Hegge, J.O.; Noble, M.A.; Lewis, D.L.; Herweijer, H.; Wolff, J.A. Progress toward a nonviral gene therapy protocol for the treatment of anemia. Hum. Gene Ther. 2007, 18, 269–285. [Google Scholar] [CrossRef]
- Le Guiner, C.; Montus, M.; Servais, L.; Cherel, Y.; Francois, V.; Thibaud, J.-L.; Wary, C.; Matot, B.; Larcher, T.; Guigand, L.; et al. Forelimb Treatment in a Large Cohort of Dystrophic Dogs Supports Delivery of a Recombinant AAV for Exon Skipping in Duchenne Patients. Mol. Ther. 2014, 22, 1923–1935. [Google Scholar] [CrossRef]
- Yasuzaki, Y.; Yamada, Y.; Fukuda, Y.; Harashima, H. Condensation of Plasmid DNA Enhances Mitochondrial Association in Skeletal Muscle Following Hydrodynamic Limb Vein Injection. Pharmaceuticals 2014, 7, 881–893. [Google Scholar] [CrossRef]
- Yasuzaki, Y.; Yamada, Y.; Kanefuji, T.; Harashima, H. Localization of exogenous DNA to mitochondria in skeletal muscle following hydrodynamic limb vein injection. J. Control. Release 2013, 172, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Le Guiner, C.; Servais, L.; Montus, M.; Larcher, T.; Fraysse, B.; Moullec, S.; Allais, M.; François, V.; Dutilleul, M.; Malerba, A.; et al. Long-term microdystrophin gene therapy is effective in a canine model of Duchenne muscular dystrophy. Nat. Commun. 2017, 8, 16105. [Google Scholar] [CrossRef] [PubMed]
- Wooddell, C.I.; Hegge, J.O.; Sebestyén, M.G.; Noble, M.; Griffin, J.B.; Pfannes, L.V.; Herweijer, H.; Hagstrom, J.E.; Braun, S.; Huss, T.; et al. Dose response in rodents and nonhuman primates after hydrodynamic limb vein delivery of naked plasmid DNA. Hum. Gene Ther. 2011, 22, 889–903. [Google Scholar] [CrossRef] [PubMed]
- Nagata, K.; Itaka, K.; Baba, M.; Uchida, S.; Ishii, T.; Kataoka, K. Muscle-targeted hydrodynamic gene introduction of insulin-like growth factor-1 using polyplex nanomicelle to treat peripheral nerve injury. J. Control. Release 2014, 183, 27–34. [Google Scholar] [CrossRef]
- Fan, Z.; Kocis, K.; Valley, R.; Howard, J.F.; Chopra, M.; Chen, Y.; An, H.; Lin, W.; Muenzer, J.; Powers, W.; et al. High-Pressure Transvenous Perfusion of the Upper Extremity in Human Muscular Dystrophy: A Safety Study with 0.9% Saline. Hum. Gene Ther. 2015, 26, 614–621. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Kocis, K.; Valley, R.; Howard, J.F.; Chopra, M.; An, H.; Lin, W.; Muenzer, J.; Powers, W. Safety and feasibility of high-pressure transvenous limb perfusion with 0.9% saline in human muscular dystrophy. Mol. Ther. 2012, 20, 456–461. [Google Scholar] [CrossRef]
- Carrasco, M.J.; Alishetty, S.; Alameh, M.-G.; Said, H.; Wright, L.; Paige, M.; Soliman, O.; Weissman, D.; Cleveland, T.E.; Grishaev, A.; et al. Ionization and structural properties of mRNA lipid nanoparticles influence expression in intramuscular and intravascular administration. Commun. Biol. 2021, 4, 956. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Tuyishime, S.; Muramatsu, H.; Kariko, K.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; Hope, M.J.; Weissman, D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release 2015, 217, 345–351. [Google Scholar] [CrossRef]
- Di, J.; Du, Z.; Wu, K.; Jin, S.; Wang, X.; Li, T.; Xu, Y. Biodistribution and Non-linear Gene Expression of mRNA LNPs Affected by Delivery Route and Particle Size. Pharm. Res. 2022, 39, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Hassett, K.J.; Benenato, K.E.; Jacquinet, E.; Lee, A.; Woods, A.; Yuzhakov, O.; Himansu, S.; Deterling, J.; Geilich, B.M.; Ketova, T.; et al. Optimization of Lipid Nanoparticles for Intramuscular Administration of mRNA Vaccines. Mol. Ther. Nucleic Acids 2019, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kanayama, N.; Fukushima, S.; Nishiyama, N.; Itaka, K.; Jang, W.-D.; Miyata, K.; Yamasaki, Y.; Chung, U.-I.; Kataoka, K. A PEG-Based Biocompatible Block Catiomer with High Buffering Capacity for the Construction of Polyplex Micelles Showing Efficient Gene Transfer toward Primary Cells. ChemMedChem 2006, 1, 439–444. [Google Scholar] [CrossRef]
- Uchida, S.; Itaka, K.; Uchida, H.; Hayakawa, K.; Ogata, T.; Ishii, T.; Fukushima, S.; Osada, K.; Kataoka, K. In Vivo Messenger RNA Introduction into the Central Nervous System Using Polyplex Nanomicelle. PLoS ONE 2013, 8, e56220. [Google Scholar] [CrossRef] [PubMed]
- Japan SLC, Inc. Laboratory Animals. Available online: http://www.jslc.co.jp/english/animals/mouse.php (accessed on 14 July 2023).
- Otto, G.P.; Rathkolb, B.; Oestereicher, M.A.; Lengger, C.J.; Moerth, C.; Micklich, K.; Fuchs, H.; Gailus-Durner, V.; Wolf, E.; de Angelis, M.H. Clinical Chemistry Reference Intervals for C57BL/6J, C57BL/6N, and C3HeB/FeJ Mice (Mus musculus). J. Am. Assoc. Lab. Anim. Sci. 2016, 55, 375–386. Available online: http://www.ncbi.nlm.nih.gov/pubmed/27423143 (accessed on 14 July 2023).
- Mazzaccara, C.; Labruna, G.; Cito, G.; Scarfò, M.; De Felice, M.; Pastore, L.; Sacchetti, L. Age-Related Reference Intervals of the Main Biochemical and Hematological Parameters in C57BL/6J, 129SV/EV and C3H/HeJ Mouse Strains. PLoS ONE 2008, 3, e3772. [Google Scholar] [CrossRef] [PubMed]
- Deuis, J.R.; Dvorakova, L.S.; Vetter, I. Methods Used to Evaluate Pain Behaviors in Rodents. Front. Mol. Neurosci. 2017, 10, 284. [Google Scholar] [CrossRef]
- Injac, R.; Perse, M.; Obermajer, N.; Djordjevic-Milic, V.; Prijatelj, M.; Djordjevic, A.; Cerar, A.; Strukelj, B. Potential hepatoprotective effects of fullerenol C60(OH)24 in doxorubicin-induced hepatotoxicity in rats with mammary carcinomas. Biomaterials 2008, 29, 3451–3460. [Google Scholar] [CrossRef] [PubMed]
- Gowda, S.; Desai, P.B.; Kulkarni, S.S.; Hull, V.V.; Math, A.A.K.; Vernekar, S.N. Markers of renal function tests. N. Am. J. Med. Sci. 2010, 2, 170–173. Available online: https://www.najms.org (accessed on 29 March 2023).
- Fukada, S.I.; Higashimoto, T.; Kaneshige, A. Differences in muscle satellite cell dynamics during muscle hypertrophy and regeneration. Skelet. Muscle 2022, 12, 17. [Google Scholar] [CrossRef]
- Hamer, P.W.; McGeachie, J.M.; Davies, M.J.; Grounds, M.D. Evans Blue Dye as an in vivo marker of myofibre damage: Optimising parameters for detecting initial myofibre membrane permeability. J. Anat. 2002, 200, 69–79. [Google Scholar] [CrossRef]
- Saria, A.; Lundberg, J.M. Evans blue fluorescence: Quantitative and morphological evaluation of vascular permeability in animal tissues. J. Neurosci. Methods 1983, 8, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Tahtinen, S.; Tong, A.-J.; Himmels, P.; Oh, J.; Paler-Martinez, A.; Kim, L.; Wichner, S.; Oei, Y.; McCarron, M.J.; Freund, E.C.; et al. IL-1 and IL-1ra are key regulators of the inflammatory response to RNA vaccines. Nat. Immunol. 2022, 23, 532–542. [Google Scholar] [CrossRef]
- Kim, J.; Eygeris, Y.; Gupta, M.; Sahay, G. Self-assembled mRNA vaccines. Adv. Drug Deliv. Rev. 2021, 170, 83–112. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.E.; Frenck, R.W., Jr.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based COVID-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef] [PubMed]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Medhurst, S.J.; Walker, K.; Bowes, M.; Kidd, B.L.; Glatt, M.; Muller, M.; Hattenberger, M.; Vaxelaire, J.; O’Reilly, T.; Wotherspoon, G.; et al. A rat model of bone cancer pain. Pain 2002, 96, 129–140. [Google Scholar] [CrossRef]
- Schött, E.; Berge, O.-G.; Ängeby-Möller, K.; Hammarström, G.; Dalsgaard, C.-J.; Brodin, E. Weight bearing as an objective measure of arthritic pain in the rat. J. Pharmacol. Toxicol. Methods 1994, 31, 79–83. [Google Scholar] [CrossRef]
- Alameh, M.-G.; Tombácz, I.; Bettini, E.; Lederer, K.; Ndeupen, S.; Sittplangkoon, C.; Wilmore, J.R.; Gaudette, B.T.; Soliman, O.Y.; Pine, M.; et al. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity 2021, 54, 2877–2892. [Google Scholar] [CrossRef]
- Cross, R. Can mRNA disrupt the drug industry? CEN Glob. Enterp. 2018, 96, 35–40. [Google Scholar] [CrossRef]
- Garber, K. mRNA pioneers refocus on therapeutics. Nat. Rev. Drug Discov. 2022, 21, 699–701. [Google Scholar] [CrossRef] [PubMed]
- Collén, A.; Bergenhem, N.; Carlsson, L.; Chien, K.R.; Hoge, S.; Gan, L.-M.; Fritsche-Danielson, R. VEGFA mRNA for regenerative treatment of heart failure. Nat. Rev. Drug Discov. 2022, 21, 79–80. [Google Scholar] [CrossRef] [PubMed]
- Lutz, J.; Lazzaro, S.; Habbeddine, M.; Schmidt, K.E.; Baumhof, P.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; Hope, M.J.; Heidenreich, R.; et al. Unmodified mRNA in LNPs constitutes a competitive technology for prophylactic vaccines. npj Vaccines 2017, 2, 29. [Google Scholar] [CrossRef] [PubMed]
- Bernard, M.-C.; Bazin, E.; Petiot, N.; Lemdani, K.; Commandeur, S.; Verdelet, C.; Margot, S.; Perkov, V.; Ripoll, M.; Garinot, M.; et al. The impact of nucleoside base modification in mRNA vaccine is influenced by the chemistry of its lipid nanoparticle delivery system. Mol. Ther. Nucleic Acids 2023, 32, 794–806. [Google Scholar] [CrossRef]








| Gene | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) |
|---|---|---|
| Il6 | TACCACTTCACAAGTCGGAGGC | CTGCAAGTGCATCATCGTTGTTC |
| Il1b | TGGACCTTCCAGGATGAGGACA | GTTCATCTCGGAGCCTGTAGTG |
| Tnf | CCACGTCGTAGCAAACCACC | TTGAGATCCATGCCGTTGGC |
| Actb | GTGACGTTGACATCCGTAAAGA | GCCGGACTCATCGTACTCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, X.; Yada, E.; Terai, Y.; Takahashi, T.; Nakanishi, H.; Tanaka, H.; Akita, H.; Itaka, K. Comprehensive Evaluation of Lipid Nanoparticles and Polyplex Nanomicelles for Muscle-Targeted mRNA Delivery. Pharmaceutics 2023, 15, 2291. https://doi.org/10.3390/pharmaceutics15092291
Du X, Yada E, Terai Y, Takahashi T, Nakanishi H, Tanaka H, Akita H, Itaka K. Comprehensive Evaluation of Lipid Nanoparticles and Polyplex Nanomicelles for Muscle-Targeted mRNA Delivery. Pharmaceutics. 2023; 15(9):2291. https://doi.org/10.3390/pharmaceutics15092291
Chicago/Turabian StyleDu, Xuan, Erica Yada, Yuki Terai, Takuya Takahashi, Hideyuki Nakanishi, Hiroki Tanaka, Hidetaka Akita, and Keiji Itaka. 2023. "Comprehensive Evaluation of Lipid Nanoparticles and Polyplex Nanomicelles for Muscle-Targeted mRNA Delivery" Pharmaceutics 15, no. 9: 2291. https://doi.org/10.3390/pharmaceutics15092291