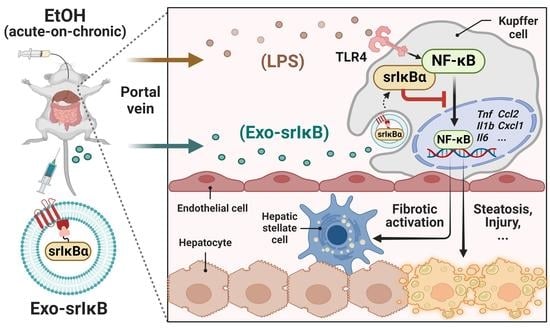
1. Introduction
Alcohol-associated liver disease (ALD) caused by chronic and excessive alcohol consumption is one of the most prevalent chronic liver diseases worldwide [
1]. Usually, ALD follows a well-defined disease spectrum starting with the alcohol-associated fatty liver (AFL) that develops into alcohol-associated steatohepatitis, fibrosis (ALF), cirrhosis (ALC), and hepatocellular carcinoma (HCC) [
2]. Chronic alcohol consumption induces gut dysbiosis and increases intestinal permeability, resulting in the hepatic influx of abnormally high amounts of pathogen-associated molecular patterns, including lipopolysaccharide (LPS), through the portal circulation [
1]. Although Kupffer cells (KCs) take up and detoxify gut-derived LPS under the homeostatic condition [
3], elevated levels of LPS by chronic alcohol consumption are sensed by toll-like receptor 4 (TLR4) on KCs and initiate alcohol-associated hepatic inflammation. In particular, TLR4-mediated activation of nuclear factor-κB (NF-κB) triggers the production of pro-inflammatory cytokines and chemokines such as tumor necrosis factor (TNF)-α, interleukin (IL)-6, pro-IL-1β, C-C motif chemokine ligand 2 (CCL2), and C-X-C motif chemokine ligand 1 (CXCL1) by KCs and accelerates the progression of ALD [
1,
4,
5]. Therefore, targeting NF-κB signaling in KCs may be a viable therapeutic approach in ALD.
The LPS-mediated NF-κB signaling activation involves the transfer of LPS by LPS-binding protein (LBP) to a cluster of differentiation 14 (CD14), followed by binding with the TLR4/myeloid differentiation factor 2 complex. Oligomerization of TLR4 triggers recruitment of downstream adaptors to myeloid differentiation primary response gene 88 (MyD88), resulting in the consequential activation of transforming growth factor β-activated kinase 1 (TAK-1) [
6]. In ALD, TAK-1 activation provokes the release of NF-κB and its nuclear translocation via IκB kinase (IKK)-mediated phosphorylation of IκB, unlike the homeostatic state in which IκB binds to NF-κB in the cytosol and restricts its nuclear translocation [
7]. It has been reported that mice deficient with LBP [
8], CD14 [
9], or TLR4 [
10] showed decreased alcohol-associated liver injury (ALI) compared to wild-type (WT) controls. The above promising results from animal experiments prompted the discovery or synthesis of therapeutic agents that suppress NF-κB activation for clinical usage in ALD. Indeed, more than 700 NF-κB inhibitors that target various stages of the NF-κB signaling pathway have been developed [
11], and some of them have commenced clinical trials for inflammatory liver diseases, including autoimmune hepatitis, non-alcoholic steatohepatitis, and chronic hepatitis C viral infection [
12]. Despite these efforts, however, there are no approved NF-κB blockers and therapeutics for ALD except liver transplantation or abstinence.
An impediment to the drug development is the successful and efficient delivery of drugs to target cells to minimize adverse effects. Nanoparticles have been highlighted for the targeted delivery of small molecules, nucleic acids, and proteins [
13]. In particular, galactose-coated nanoparticles target hepatocytes (HEPs), which exclusively express the asialoglycoprotein receptor [
14]. However, it has been well known that the reticuloendothelial system-mediated clearance limits the target cell delivery of nanoparticles [
15]. Therefore, instead, exosomes have attracted attention as a novel drug carrier system. Exosomes are extracellular vesicles that are between 40 and 120 nm in size and can be generated and secreted by the various cell types in our body. The biological function of exosomes is to mediate intercellular communication by delivering diverse cargo, including nucleic acids, proteins, and lipids [
16]. Exosome-based drug delivery provides many advantages since it has a high capacity to store drugs, overcome biological barriers, and target specificity [
17]. Recently, a novel, optogenetically engineered exosome technology called ‘exosomes for protein loading via optically reversible protein–protein interactions (EXPLOR)’ has been developed that enhances the biological compatibility and production efficiency of exosomes [
18]. In addition, by utilizing EXPLORs, Yim et al. engineered exosomes to load IκB protein without IKK phosphorylation sites called ‘super-repressor IκB’ (Exo-srIκB) that has a prolonged half-life and sustained inhibition of NF-κB [
18]. Follow-up studies with Exo-srIκB have demonstrated that in vivo delivery of Exo-srIκB attenuates sepsis-associated organ damage and mortality, inflammatory responses related to preterm birth, and ischemia-reperfusion kidney injury in mice [
19,
20,
21]. In addition, in vivo tracing of the biodistribution of zirconium-89 (
89Zr)-labeled Exo-srIκB in mice revealed a predominant delivery of Exo-srIκB to the liver after intravenous injection [
22].
Based on the above findings, here, we aimed to assess the protective effects of Exo-srIκB on ALI. Since there is no approved drug for ALD except abstinence or liver transplantation [
1], the development of novel therapies is urgently needed. In this aspect, our findings may advance the field of ALD by providing Exo-srIκB as a useful therapeutic agent.
2. Materials and Methods
2.1. Mice
C57BL/6J WT male mice (Jackson Laboratory, Bar Harbor, ME, USA) aged 8 to 10 weeks (about 25 g of body weight) were used for in vivo experiments. All in vivo studies were approved by the Institutional Animal Care and Use Committee of the Korea Advanced Institute of Science and Technology (KAIST) (Approved No. KA2021-071). Mice were maintained in a specific pathogen-free facility at KAIST with a standard 12 h light and dark cycle. There was no bias for the location, humidity, and temperature of cages. Mice were acclimatized with feeding location for at least 1 week and with Lieber–DeCarli ethanol (EtOH) diet (Dyets Inc., Bethlehem, PA, USA) for 5 days. After that, the percentage of EtOH in the diet was gradually increased (1 to 5%) for another 5 days. After a total of 17 days of acclimatization periods, mice were fed with 5% EtOH-containing diet for 10 days [
23]. On day 10, mice were randomly divided into each group (
n = 6 per group) based on body weight. Therefore, there was no difference in body weight before exosome treatment among experimental groups. The sample size was decided based on previous experiences to obtain meaningful data and the availability of mouse facility, and there were no exclusions.
For the single-injection experiment, mice were randomly divided into control exosome generated from un-engineered wild-type Expi293F cells (Exo-Naïve)- or Exo-srIκB-injected group (
n = 6 per group) on day 10, and exosomes (5.0 × 10
10 particles per mouse) were intravenously administered via tail vein. After 1 h of exosome injection, 4 g kg
−1 of 40% EtOH was orally delivered to each mouse and they were sacrificed after 6 h of EtOH gavage [
23]. For experiments of 3 consecutive days of injection, mice were randomly assigned in Exo-Naïve (10
9 particle per mouse)-, low doses of Exo-srIκB (10
8 particles per mouse)-, or high doses of Exo-srIκB (10
9 particles per mouse)-injected group (
n = 6 per group) on day 10. Indicated doses of exosomes were intravenously delivered for 3 consecutive days with 24-hour intervals. After 6 h of last exosome injection, 4 g kg
−1 of 40% EtOH was orally administered to each mouse. Doses of exosomes used in the present study were determined based on the previous experiments [
19,
20,
21]. Since many hepatic cells underwent apoptosis after acute high doses of EtOH gavage and caused injury, we isolated hepatic cells after 1 h of EtOH gavage for analysis. However, mice were sacrificed after 6 h of EtOH gavage similar to single-injection experiment. Researchers were not blinded for the animal experiments.
2.2. Exosomes Production
The exosome production process was previously described [
19,
24]. In brief, Expi293F producing cells were incubated for 4 days in a wave culture system, and cells were exposed to blue-light illumination for target protein loading and exosome production. After that, the culture medium was harvested and centrifuged at 2000×
g for 10 min to remove cells and debris. A 0.22 µm of polyethersulfone filter was used to remove large particles. Next, the exosome was purified through the ultrafiltration and diafiltration process for the concentrated harvested culture medium and buffer exchange. Then, exosomes were purified through anionic and multi-modal resin chromatography. Finally, a formulation and sterilization filter process was performed.
2.3. Transmission Electron Microscopy
The morphology of extracellular vesicles (EVs) was characterized by transmission electron microscope (TEM). First, EVs were allowed to absorb onto carbon-coated copper grids (Electron Microscopy Sciences, Hatfield, PA, USA) for 15 sec. After removing excess liquid, samples were negatively stained with 2% uranyl acetate (Electron Microscopy Sciences, Hatfield, PA, USA). TEM images were obtained with a Tecnai G2 Retrofit electron microscope (FEI Company, Hillsboro, OR, USA) operating at 200 kV.
2.4. Nanoparticle Tracking Analysis
Particle number and size distribution of EVs were measured by nanoparticle tracking analysis (NTA) using NS300 (Malvern Panalytical a spectris company, Malvern, UK). According to the manufacturer’s manual, samples were diluted (1:100 to 1:10,000) in particle-free saline to an acceptable concentration. Samples were analyzed under constant flow conditions at 25 °C, a camera level of 15, and detect threshold of 3. Concentrations of EVs were measured on the basis of counts of 20 to 100 particles per frame.
2.5. Next-Generation Sequencing Analyses
Single-cell RNA sequencing (scRNA-seq) analysis of normal mouse liver tissues [
25] can be explored on the Tabula Muris Senis website (
https://tabula-muris-senis.ds.czbiohub.org/ (accessed on 12 December 2022)) or found in NCBI Gene Expression Omnibus under accession number GSE132042. The bulk RNA-seq of a vehicle or LPS-treated mouse primary KCs [
26] is publicly available in NCBI Gene Expression Omnibus under accession number GSE86397. Kyoto encyclopedia of genes and genomes (KEGG) pathway and gene ontology analyses were performed with Database for Annotation, Visualization and Integrated Discovery (DAVID) (
https://david.ncifcrf.gov/ (accessed on 21 December 2022).) [
27]. scRNA-seq analysis of human liver specimens [
28] is publicly available in NCBI Gene Expression Omnibus under accession number GSE136103.
2.6. Isolation of Mouse Primary Hepatic Cells
HEPs, hepatic stellate cells (HSCs), KCs, and liver mononuclear cells (MNCs) were isolated from C57BL/6J WT male mice by differential centrifugation on an Opti-Prep (Sigma-Aldrich, St. Louis, MO, USA) or Percoll (Sigma-Aldrich, St. Louis, MO, USA) as previously described [
29]. In brief, a two-step collagenase liver perfusion was performed by portal vein cannulation. The liver was first perfused with EGTA solution (5.4 mM KCl, 0.44 mM KH
2PO
4, 140 mM NaCl, 0.34 mM Na
2HPO
4, 0.5 mM EGTA, 25 mM Tricine, and pH 7.2), followed by the collagenase solution (0.075% collagenase type I (Worthington, Columbus, OH, USA) in Hank’s balanced salt solution (HBSS) with 0.02% DNase I). After complete circulation, the liver was extracted and further digested in the digestion buffer (0.009% of collagenase type I in HBSS with 0.02% of DNase I) at 37 °C in a shaking incubator (90 rpm, 20 min). The digested liver was then filtered through a 70 μm cell strainer to remove undigested debris and connective tissues. To isolate HEP, filtered cell suspension was centrifuged at 50×
g for 5 min and further purified by 50% Percoll gradient solution. To isolate hepatic non-parenchymal cells, the supernatant from the first centrifugation was obtained and centrifuged at 650×
g for 10 min. From the whole non-parenchymal cell pellet, HSCs and KCs were isolated by centrifugation at 1800×
g, 4 °C, for 17 min with 11.5 % and 20 % Opti-prep gradient solution, respectively. Cells obtained were further washed in HBSS before use. For isolation of liver MNCs, the non-parenchymal cell pellet was suspended in 40% Percoll gradient solution and centrifuged at 1800×
g for 20 min. The supernatant, containing debris, was carefully removed and liver MNCs were resuspended in saline following the lysis of red blood cells.
2.7. DiI Labeling of Exo-srIκB
Isolated and purified Exo-srIκB (1.0 × 1012 particles) was incubated with 10 μL of DiI at 37 °C for 30 min protected from light. After that, 1 × filtered phosphate-buffered saline was added to up to 4 mL and moved to Amicon® Ultra-4 centrifugal filter unit (Merck, Rahway, NJ, USA). Solutions were eluted by centrifugation at 3200× g, 4 °C, for 15 min until the residual volume was approximately 200 μL. Exo-srIκB was directly treated to the culture media of KCs.
2.8. Quantitative Polymerase Chain Reaction
Total RNA was extracted from liver tissues or isolated hepatic cells using TRIzol
® (Thermo Fisher Scientific, Waltham, MA, USA) and NanoDrop™ Lite (Thermo Fisher Scientific, Waltham, MA, USA) was used to measure the quantity and quality of isolated RNA. A sample had A260/A280 ratio over 1.8 which was used. The cDNA then synthesized from the extracted RNA with ReverTra Ace
® qPCR RT Master Mix with gDNA Remover (Toyobo, Osaka, Japan). A quantitative real-time polymerase chain reaction (qRT-PCR) was performed with SYBR Green Real-time PCR Master Mix (Toyobo, Osaka, Japan). All the samples were duplicated in qRT-PCR analyses. The mRNA expression levels of the genes of interest were normalized by
18s rRNA expression levels. The primer sequences used in qRT-PCR analyses are listed in
Table S1.
2.9. Histological Analyses
For consistency of sample comparison, similar regions of the left and medial lobes of mouse liver were used for histological analyses. Liver tissues were fixed with 10% neutral buffer formalin (Sigma-Aldrich, St. Louis, MO, USA) overnight at room temperature. After deparaffinization and rehydration, the paraffin-embedded tissues were sliced at 4 μm thickness and stained with hematoxylin (Sigma-Aldrich, St. Louis, MO, USA) and eosin (Sigma-Aldrich, St. Louis, MO, USA) solution. For Oil Red O staining, formalin-fixed liver tissues were embedded in frozen section compound (Leica Biosystems, Wetzlar, DE) and sliced at 10 μm thickness before use. Stained images were captured using light microscopy (Olympus BX51) and analyzed with DP2-BSW software.
Paraffin-embedded liver tissues sectioned at 4 μm thickness were used for immunostaining. Following deparaffinization and rehydration, tissue samples were immersed in 10 mM citrate buffer (pH 6.0) and microwaved for 5 min for antigen retrieval. Tissues were first blocked with 10% donkey serum for 1 h at room temperature, followed by an overnight incubation with C-type lectin domain family 4 member F (CLEC4F) (R&D Systems, MN, USA) or myeloperoxidase (MPO) (Abcam, Cambridge, UK) antibodies (1:50 to 1:200 in saline containing 0.1% Tween-20) at 4 °C. For CLEC4F immunofluorescence staining, sections were incubated with Alexa Fluor® 594-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA, USA) for 1 h at room temperature and covered with DAPI mounting solution (Abcam, Cambridge, UK). For MPO immunohistochemistry analysis, samples were incubated with either anti-Rabbit IgG (Vector Laboratories, Newark, CA, USA) or for 1 h at room temperature and developed with 3.3′-diaminobenzidine (DAB) substrate kit (Vector Laboratories, Newark, CA, USA), followed by Balsam (Sigma-Aldrich, St. Louis, MO, USA) covering. The terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay (Abcam, Cambridge, UK) was performed according to the manufacturer’s instructions. Images were captured using Olympus BX51 microscope equipped with a CCD camera (Olympus, Tokyo, Japan) and analyzed with DP2-BSW.
2.10. Biochemical Analysis
For hepatic triglyceride (TG) measurement, hepatic lipids were extracted from about 20 to 30 mg of frozen liver tissues using chloroform/methanol (2:1 ratio) solution as previously described [
30]. Lyophilized hepatic lipids were then resuspended to 5% bovine serum albumin (BSA)-saline. The VetTest Chemistry analyzer (IDEXX Laboratories, Westbrook, ME, USA) was used to measure the hepatic TG levels and serum levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), TG, and total cholesterol (TC), according to the manufacturer’s protocols.
2.11. Western Blotting
Expi293F producing cells were lysed in RIPA buffer containing Halt™ Protease and Phosphatase Inhibitor Cocktail (100×) (Thermo Fisher Scientific, Waltham, MA, USA). Lysates were centrifuged (12,000 rpm) at 4 °C for 20 min, and the supernatant was used for immunoblotting. Exosome was lysed in 4X Laemmli sample buffer (Bio-RAD, Hercules, CA, USA). Cell lysate protein and exosomes were boiled for 5 min at 100 °C. Samples were run in 10% SDS/polyacrylamide gel electrophoresis (PAGE) gel and transferred onto nitrocellulose membrane. The membranes were blocked by 5% skim milk in Tris-buffered saline containing 0.1% Tween-20 (TBS-T) for 1 h at RT. The membrane was incubated with primary antibodies against srIκB, CRY2 (customized antibody, Abclon, Seoul, Korea), CD9, CD81 (SBI, Tokyo, Japan), TSG101, alix, GM130, calnexin (Abcam, Cambridge, UK), Lamin B1, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Santa Cruz Biotechnology, Dallas, TX, USA), and prohibitin (NOVUSBIO, Centennial, CO, USA) at 4 °C overnight. After incubation with specific secondary antibodies, blots were developed using Clarity and Clarity Max ECL Western Blotting Substrates and imaged with the ChemiDoc imager (Bio-Rad, Hercules, CA, USA).
Frozen liver tissues were first subjected to nuclear extraction using the nuclear and cytoplasmic extraction kit (Thermo Fisher Scientific, Waltham, MA, USA). Proteins extracted from the nuclear compartments (to 30 μg) were first separated in a 10% SDS-PAGE, and then transferred onto a nitrocellulose membrane. After the transfer, the membrane was blocked with 5% skim milk solution for 1 h at room temperature, followed by an overnight incubation with NF-κB or lamin B1 (Abcam, Cambridge, UK) antibodies (1:1000 to 1:2000 in saline containing 0.1% Tween-20) at 4 °C. On the next day, membranes were washed and incubated with anti-rabbit secondary antibodies (Cell Signaling Technology, Danvers, MA, USA) for 1 hour at room temperature. Immunoreactive bands were detected by SuperSignal™ West Femto PLUS Chemiluminescent substrate (Thermo Fisher Scientific, Waltham, MA, USA) and captured by ImageQuant™ LAS 4000 (GE Healthcare, Chicago, IL, USA). Nuclear protein levels were normalized to the expression of lamin B1 for each sample. Densitometry analysis was performed with ImageJ (National Institute of Health, Bethesda, MD, USA).
2.12. Flow Cytometry Analyses
Isolated liver MNCs were stained with Live/Dead Fixable Aqua Dead Cell Stain Kit (Invitrogen, MA, Waltham, USA) and incubated at room temperature for 15 min protected from light. After washing with flow cytometry buffer solution (0.5% BSA, 0.955% Dulbecco’s phosphate buffered saline, and 0.05% sodium azide) by centrifugation at 350× g, 4 °C, for 5 min, samples were labeled with anti-mouse CD16/CD232 (BD Biosciences, Franklin Lakes, NJ, USA) and fluorescence-conjugated eFluor450 anti-mouse CD45 (Invitrogen, Waltham, MA, USA), peridinin-chlorophyll-protein (PerCP)-cyanine (Cy)5.5 anti-mouse CD45 (BD Biosciences, Franklin Lakes, NJ, USA), brilliant violet (BV)786 anti-mouse CD3e (BD Biosciences, Franklin Lakes, NJ, USA), PerCP-Cy5.5 anti-mouse CD4 (BD Biosciences, Franklin Lakes, NJ, USA), allophycocyanin (APC) anti-mouse CD8a (BD Biosciences, Franklin Lakes, NJ, USA), phycoerythrin (PE) anti-mouse NK1.1 (BD Biosciences, Franklin Lakes, NJ, USA), fluorescein isothiocyanate (FITC) anti-mouse F4/80 (Invitrogen, Waltham, MA, USA), PE-Cy7 anti-mouse F4/80 (Invitrogen, Waltham, MA, USA), PE anti-mouse Ly6C (BD Biosciences, Franklin Lakes, NJ, USA), PerCP-Cy5.5 anti-mouse Ly6G (BD Biosciences, Franklin Lakes, NJ, USA), APC-Cy7 anti-mouse CD11b (BD Biosciences, Franklin Lakes, NJ, USA), PE-CF594 anti-mouse Siglec-F (BD Biosciences, Franklin Lakes, NJ, USA), PE-CF594 anti-mouse Ly6G (BD Biosciences, Franklin Lakes, NJ, USA), PE anti-mouse C-type lectin-like receptor 2 (CLEC2) (BioLegend, San Diego, CA, USA), Alexa Fluor® 647 anti-mouse T-cell immunoglobulin mucin-4 (TIM4) (BioLegend, San Diego, CA, USA), and PE-Cy7 anti-mouse CD146 (BioLegend, San Diego, CA, USA) antibodies. After washing with flow cytometry buffer solution by centrifugation at 350× g, 4 °C, for 5 min, stained cells were read with LSRFortessa™ X-20 (BD Biosciences, Franklin Lakes, NJ, USA) and analyzed with FlowJo™ v10 software (BD Biosciences, Franklin Lakes, NJ, USA).
2.13. Enzyme-Linked Immunosorbent Assay (ELISA)
Mouse IL-6 (#M6000B), IL-1β (#MLB00C), and TNF-α (#MTA00B) Quantikine ELISA kits (R&D Systems, Minneapolis, MN, USA) were used according to the manufacturer’s instructions. Microplates were read by an iMarkTM microplate absorbance reader (Bio-Rad, CA, USA).
2.14. Statistical Analyses
All statistical analyses in this study were conducted with Prism version 8.0 (GraphPad Software, San Diego, CA, USA). Statistical significance was examined by a two-tailed Student’s t-test between the two groups or a one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test among three or more groups. For RNA-seq, an adjusted p value was used. p < 0.05 was thought to be statistically meaningful.
4. Discussions
Chronic alcohol consumption results in an upsurge in intestinal permeability, which leads to the hepatic translocation of endotoxins (e.g., LPS) through the portal vein. The continuous influx of LPS instigates alcohol-associated hepatic inflammation by activating TLR4/NF-κB signaling pathways in KCs. In particular, NF-κB activation in KCs promotes the production of inflammatory cytokines (e.g., TNF-α, IL-6, and IL-1β) and chemokines (e.g., CCL2 and CXCL1) to induce death of HEPs and recruitment of inflammatory monocytes and neutrophils [
1]. Although there have been many efforts to abate these processes, there is still no approved drug for ALD. Since liver damage could not be reversed in the late stage of ALD such as ALC or HCC [
2], there is an urgent need for a new therapeutic agent, especially targeting early-stage ALD. In the present study, we provide several lines of evidence for the efficient preventive effect of Exo-srIκB against ALI, AFL, and ALF in mice. In particular, we reveal that the beneficial effects of Exo-srIκB are mediated primarily by limiting the inflammatory activation of KCs. These findings imply that exosome-based targeting inflammatory KCs may have therapeutic potential in ALD. In addition, our results expand the prior research demonstrating the protective role of Exo-srIκB in the inflammatory responses of sepsis [
19], preterm birth [
20], and ischemia-reperfusion kidney injury [
21], suggesting the possible application of Exo-srIκB to broad inflammatory diseases.
Previous studies with LBP [
8], CD14 [
9], or TLR4 [
10] knockout (KO) mice reported a crucial role of NF-κB signaling pathway in the development of ALD. Interestingly, the ALI is not reduced in MyD88 KO mice, indicating alcohol-specific MyD88-independent activation of NF-κB [
35]. However, since the above studies used whole-body KO mice, identifying the cell type for drug targeting is an arduous task. Therefore, despite promising results in animal experiments, there are no clinical trials conducted with NF-κB inhibitors in ALD. We found highly enriched gene expression levels of exosome uptake processes in KCs among various hepatic cells by analyzing scRNA-seq data [
32]. In addition, inflammatory gene expression levels were most dramatically suppressed in KCs after three consecutive days of Exo-srIκB injection with attenuated ALI. These data suggest that targeting NF-κB signaling pathways in KCs might be an effective approach in ALD.
KCs are heterogenous populations depending on their origins. Originally, KCs are derived from the yolk sac and reside in the liver sinusoid. However, chronic liver injury in (non-)alcoholic liver diseases triggers apoptosis of emKCs and their replacement by bmKCs, which are primarily regulated by liver X receptor-α signaling pathways [
33,
34]. Interestingly, we showed that systemic administration of Exo-srIκB prevented the pro-inflammatory activation of KCs without affecting the frequencies of emKCs and bmKCs in mice with ALD. In addition, the morphological changes in CLEC4F
+ KCs of Exo-Naïve-treated EtOH-fed mice in the midzonal area disappeared by Exo-srIκB treatment. Moreover, all of the pathologic findings, including lipid accumulation, apoptosis, and infiltration of MPO
+ neutrophils, were found in the hepatic midzonal area of Exo-Naïve-treated mice. These results may indicate that there might be a functional heterogeneity in KCs depending on their locations across the hepatic lobules in ALD; perivenous KCs undergo apoptosis, midzonal KCs provoke inflammation, and periportal KCs limit endotoxin levels by phagocytosis [
36,
37]. It would be interesting to define the functional and spatial heterogeneity of KCs in detail by utilizing spatial multi-omics technology for more precise targeting of inflammatory KCs in ALD.
Recent advances in nanotechnology enable the development of nanomedicine for small-molecule therapeutics. However, this approach needs a high dose of the drug because of the low target specificity, resulting in severe adverse effects [
13]. In our animal experiments, we showed a similar extent of preventative effects in low (10
8 particles) and high (10
9 particles) doses of Exo-srIκB against acute-on-chronic ALI without unexpected rise of ALT in serum. These results indicate the high potency and efficiency of Exo-srIκB in ALD, consistent with the previous finding that demonstrates liver-oriented biodistribution of Exo-srIκB [
22]. In addition, although the number of analyzed patients was small, we detected enhanced expression levels of genes related to exosome uptake processes in
MARCO+ resident KCs of patients with ALC compared to those of the healthy controls. Based on the above results, favorable outcomes can be expected from clinical trials of Exo-srIκB in patients with ALD.
Our study has some limitations. First, since the exosomes used in the present study originated from human cell lines, further study is needed to elucidate the mechanisms of action of Exo-srIκB in detail to minimize the unexpected adverse effects. In addition, we detected a significant decrease in the hepatic frequencies of NK cells after multiple injections of Exo-srIκB in mice with ALD. NK cells are innate immune cells that play important roles in protecting against viral hepatitis and liver fibrosis by killing virus-infected HEPs or fibrotic HSCs [
29,
38]. Even though Exo-srIκB treatment decreased expression levels of genes related to fibrotic activation in HSCs, the usage of Exo-srIκB in late-stage ALD should be performed with caution.
In conclusion, the present study demonstrates the protective role of Exo-srIκB in ALI, especially in suppressing inflammatory activation of KCs in the animal model. Our study has a particular significance in that it suggests a novel exosome-based therapeutic paradigm in ALD.