Oleyl Conjugated Histidine-Arginine Cell-Penetrating Peptides as Promising Agents for siRNA Delivery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis and Purification of Oleyl Conjugated Peptides
2.2. Complex Formation of Oleyl Conjugated Peptides and siRNA
2.3. Cell Culture
2.4. In Vitro Cytotoxicity Assay
2.5. Dynamic Light Scattering
2.6. Cellular Internalization of siRNA
2.7. Cellular Uptake Study Using Confocal Microscopy
2.8. Gel Shifting Assay
2.9. Protection of siRNA against Enzymatic Degradation
2.10. Protein Silencing Effect of siRNA (Western Blot)
2.11. Statistical Analysis
3. Results and Discussion
3.1. Synthesis and Characterization of the Oleyl-Conjugated Histidine-Arginine-Containing Peptide
3.2. Cytotoxicity
3.3. Characterization of Peptide-siRNA Complexes Using Dynamic Light Scattering
3.3.1. Zeta Potential
3.3.2. Particle Size
3.4. Cellular Internalization of Alexa Fluor Labeled siRNA Using Flow Cytometry
3.5. Cellular Internalization Using Confocal Microscopy
3.6. Binding Affinity of the Peptides with siRNA Using Gel Electrophoresis
3.7. Serum Stability of Peptide-siRNA Complexes
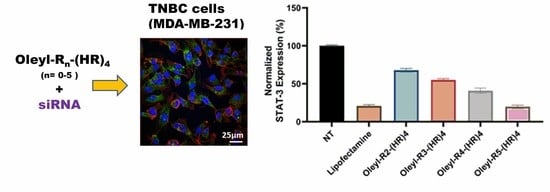
3.8. STAT-3 Silencing Using Oleyl Conjugated Peptides/siRNA Complexes in MDA-MB-231 Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Caplen, N.J.; Parrish, S.; Imani, F.; Fire, A.; Morgan, R.A. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA 2001, 98, 9742–9747. [Google Scholar] [CrossRef] [Green Version]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef]
- Sajid, M.I.; Moazzam, M.; Kato, S.; Yeseom Cho, K.; Tiwari, R.K. Overcoming barriers for siRNA therapeutics: From bench to bedside. Pharmaceuticals 2020, 13, 294. [Google Scholar] [CrossRef]
- Subhan, M.A.; Torchilin, V. siRNA based drug design, quality, delivery and clinical translation. Nanomed. Nanotechnol. Biol. Med. 2020, 29, 102239. [Google Scholar] [CrossRef]
- Park, J.W.; Bae, K.H.; Kim, C.; Park, T.G. Clustered magnetite nanocrystals cross-linked with PEI for efficient siRNA delivery. Biomacromolecules 2011, 12, 457–465. [Google Scholar] [CrossRef]
- Liu, G.; Xie, J.; Zhang, F.; Wang, Z.; Luo, K.; Zhu, L.; Quan, Q.; Niu, G.; Lee, S.; Ai, H. N-Alkyl-PEI-functionalized iron oxide nanoclusters for efficient siRNA delivery. Small 2011, 7, 2742–2749. [Google Scholar] [CrossRef] [Green Version]
- Rozema, D.B.; Lewis, D.L.; Wakefield, D.H.; Wong, S.C.; Klein, J.J.; Roesch, P.L.; Bertin, S.L.; Reppen, T.W.; Chu, Q.; Blokhin, A.V. Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc. Natl. Acad. Sci. USA 2007, 104, 12982–12987. [Google Scholar] [CrossRef] [Green Version]
- Nedra Karunaratne, D.; Jafari, M.; Udayana Ranatunga, R.; Siriwardhana, A. Natural carriers for siRNA delivery. Curr. Pharm. Des. 2015, 21, 4529–4540. [Google Scholar] [CrossRef]
- Pan, J.; Mendes, L.P.; Yao, M.; Filipczak, N.; Garai, S.; Thakur, G.A.; Sarisozen, C.; Torchilin, V.P. Polyamidoamine dendrimers-based nanomedicine for combination therapy with siRNA and chemotherapeutics to overcome multidrug resistance. Eur. J. Pharm. Biopharm. 2019, 136, 18–28. [Google Scholar] [CrossRef]
- Wu, J.; Huang, W.; He, Z. Dendrimers as carriers for siRNA delivery and gene silencing: A review. Sci. World J. 2013, 630654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.; Yang, S.-j.; Wang, J.-c.; Yang, L.-j.; Xu, Z.-z.; Yang, T.; Liu, X.-y.; Zhang, Q. Sequential treatment of drug-resistant tumors with RGD-modified liposomes containing siRNA or doxorubicin. Eur. J. Pharm. Biopharm. 2010, 76, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Chien, P.-Y.; Wang, J.; Carbonaro, D.; Lei, S.; Miller, B.; Sheikh, S.; Ali, S.M.; Ahmad, M.U.; Ahmad, I. Novel cationic cardiolipin analogue-based liposome for efficient DNA and small interfering RNA delivery in vitro and in vivo. Cancer Gene Ther. 2005, 12, 321–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroeder, A.; Levins, C.G.; Cortez, C.; Langer, R.; Anderson, D.G. Lipid-based nanotherapeutics for siRNA delivery. J. Intern. Med. 2010, 267, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.E. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: From concept to clinic. Mol. Pharm. 2009, 6, 659–668. [Google Scholar] [CrossRef]
- Lächelt, U.; Wagner, E. Nucleic acid therapeutics using polyplexes: A journey of 50 years (and beyond). Chem. Rev. 2015, 115, 11043–11078. [Google Scholar] [CrossRef]
- Semple, S.C.; Akinc, A.; Chen, J.; Sandhu, A.P.; Mui, B.L.; Cho, C.K.; Sah, D.W.; Stebbing, D.; Crosley, E.J.; Yaworski, E. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010, 28, 172–176. [Google Scholar] [CrossRef]
- Suzuki, Y.; Ishihara, H. Difference in the lipid nanoparticle technology employed in three approved siRNA (Patisiran) and mRNA (COVID-19 vaccine) drugs. Drug Metab. Pharmacokinet. 2021, 41, 100424. [Google Scholar] [CrossRef]
- Winter, E.; Pizzol, C.D.; Locatelli, C.; Crezkynski-Pasa, T.B. Development and evaluation of lipid nanoparticles for drug delivery: Study of toxicity in vitro and in vivo. J. Nanosci. Nanotechnol. 2016, 16, 1321–1330. [Google Scholar] [CrossRef]
- Fonseca-Gomes, J.; Loureiro, J.A.; Tanqueiro, S.R.; Mouro, F.M.; Ruivo, P.; Carvalho, T.; Sebastião, A.M.; Diógenes, M.J.; Pereira, M.C. In vivo bio-distribution and toxicity evaluation of polymeric and lipid-based nanoparticles: A potential approach for chronic diseases treatment. Int. J. Nanomed. 2020, 15, 8609. [Google Scholar] [CrossRef]
- Raucher, D.; Ryu, J.S. Cell-penetrating peptides: Strategies for anticancer treatment. Trends Mol. Med. 2015, 21, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Guidotti, G.; Brambilla, L.; Rossi, D. Cell-penetrating peptides: From basic research to clinics. Trends Pharmacol. Sci. 2017, 38, 406–424. [Google Scholar] [CrossRef] [PubMed]
- Lehto, T.; Ezzat, K.; Wood, M.J.; Andaloussi, S.E. Peptides for nucleic acid delivery. Adv. Drug Deliv. Rev. 2016, 106, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Moschos, S.A.; Jones, S.W.; Perry, M.M.; Williams, A.E.; Erjefalt, J.S.; Turner, J.J.; Barnes, P.J.; Sproat, B.S.; Gait, M.J.; Lindsay, M.A. Lung delivery studies using siRNA conjugated to TAT(48-60) and penetratin reveal peptide induced reduction in gene expression and induction of innate immunity. Bioconjug. Chem. 2007, 18, 1450–1459. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.H.; Hou, Y.W.; Lee, H.J. An intracellular delivery method for siRNA by an arginine-rich peptide. J. Biochem. Biophys. Methods 2007, 70, 579–586. [Google Scholar] [CrossRef]
- Kim, S.W.; Kim, N.Y.; Choi, Y.B.; Park, S.H.; Yang, J.M.; Shin, S. RNA interference in vitro and in vivo using an arginine peptide/siRNA complex system. J. Control. Release 2010, 143, 335–343. [Google Scholar] [CrossRef]
- El-Sayed, N.S.; Miyake, T.; Shirazi, A.N.; Park, S.E.; Clark, J.; Buchholz, S.; Parang, K.; Tiwari, R. Design, synthesis, and evaluation of homochiral peptides containing arginine and histidine as molecular transporters. Molecules 2018, 23, 1590. [Google Scholar] [CrossRef] [Green Version]
- Mozaffari, S.; Bousoik, E.; Amirrad, F.; Lamboy, R.; Coyle, M.; Hall, R.; Alasmari, A.; Mahdipoor, P.; Parang, K.; Montazeri Aliabadi, H. Amphiphilic peptides for efficient siRNA delivery. Polymers 2019, 11, 703. [Google Scholar] [CrossRef] [Green Version]
- Freire, J.M.; de Figueiredo, I.R.; Valle, J.; Veiga, A.S.; Andreu, D.; Enguita, F.J.; Castanho, M.A. siRNA-cell-penetrating peptides complexes as a combinatorial therapy against chronic myeloid leukemia using BV173 cell line as model. J. Control. Release 2017, 245, 127–136. [Google Scholar] [CrossRef]
- Tanaka, K.; Kanazawa, T.; Ogawa, T.; Takashima, Y.; Fukuda, T.; Okada, H. Disulfide crosslinked stearoyl carrier peptides containing arginine and histidine enhance siRNA uptake and gene silencing. Int. J. Pharm. 2010, 398, 219–224. [Google Scholar] [CrossRef]
- Biswas, A.; Chakraborty, K.; Dutta, C.; Mukherjee, S.; Gayen, P.; Jan, S.; Mallick, A.M.; Bhattacharyya, D.; Sinha Roy, R. Engineered histidine-enriched facial Lipopeptides for enhanced intracellular delivery of functional siRNA to triple negative breast Cancer cells. ACS Appl. Mater. Interfaces 2019, 11, 4719–4736. [Google Scholar] [CrossRef] [PubMed]
- Damen, M.; Aarbiou, J.; van Dongen, S.F.; Buijs-Offerman, R.M.; Spijkers, P.P.; van den Heuvel, M.; Kvashnina, K.; Nolte, R.J.; Scholte, B.J.; Feiters, M.C. Delivery of DNA and siRNA by novel gemini-like amphiphilic peptides. J. Control. Release 2010, 145, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Alshamsan, A.; Haddadi, A.; Incani, V.; Samuel, J.; Lavasanifar, A.; Uludag, H. Formulation and delivery of siRNA by oleic acid and stearic acid modified polyethylenimine. Mol. Pharm. 2009, 6, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Futaki, S.; Ohashi, W.; Suzuki, T.; Niwa, M.; Tanaka, S.; Ueda, K.; Harashima, H.; Sugiura, Y. Stearylated arginine-rich peptides: A new class of transfection systems. Bioconjugate Chem. 2001, 12, 1005–1011. [Google Scholar] [CrossRef]
- Sharma, M.; El-Sayed, N.S.; Do, H.; Parang, K.; Tiwari, R.K.; Aliabadi, H.M. Tumor-targeted delivery of siRNA using fatty acyl-CGKRK peptide conjugates. Sci. Rep. 2017, 7, 6093. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Yu, B.; Ren, W.; Mo, X.; Zhou, C.; He, H.; Jia, H.; Wang, L.; Jacob, S.T.; Lee, R.J. Enhanced hepatic delivery of siRNA and microRNA using oleic acid based lipid nanoparticle formulations. J. Control. Release 2013, 172, 690–698. [Google Scholar] [CrossRef]
- Mandal, D.; Mohammed, E.H.M.; Lohan, S.; Mandipoor, P.; Baradaran, D.; Tiwari, R.K.; Parang, K.; Aliabadi, H.M. Redox-Responsive Disulfide Cyclic Peptides: A New Strategy for siRNA Delivery. Mol. Pharm. 2022. [Google Scholar] [CrossRef]
- Aliabadi, H.M.; KC, R.B.; Bousoik, E.; Hall, R.; Barbarino, A.; Thapa, B.; Coyle, M.; Mahdipoor, P.; Uludağ, H. A systematic comparison of lipopolymers for siRNA delivery to multiple breast cancer cell lines: In vitro studies. Acta Biomater. 2020, 102, 351–366. [Google Scholar] [CrossRef]
- Aliabadi, H.M.; Parang, K.; Hall, R.; Alasmari, A. Peptide/Lipid-Associated Nucleic Acids (Plana) for Nucleic Acid Delivery. U.S. Patent US20210246019A1, 12 August 2021. [Google Scholar]
- Do, H.; Sharma, M.; El-Sayed, N.S.; Mahdipoor, P.; Bousoik, E.; Parang, K.; Montazeri Aliabadi, H. Difatty acyl-conjugated linear and cyclic peptides for siRNA delivery. ACS Omega 2017, 2, 6939–6957. [Google Scholar] [CrossRef]
- Connerty, P.; Moles, E.; de Bock, C.E.; Jayatilleke, N.; Smith, J.L.; Meshinchi, S.; Mayoh, C.; Kavallaris, M.; Lock, R.B. Development of siRNA-Loaded Lipid Nanoparticles Targeting Long Non-Coding RNA LINC01257 as a Novel and Safe Therapeutic Approach for t (8; 21) Pediatric Acute Myeloid Leukemia. Pharmaceutics 2021, 13, 1681. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Jove, R. The STATs of cancer—new molecular targets come of age. Nat. Rev. Cancer 2004, 4, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Tai, W.; Gao, X. Functional peptides for siRNA delivery. Adv. Drug Deliv. Rev. 2017, 110, 157–168. [Google Scholar] [CrossRef] [Green Version]
- Futaki, S.; Suzuki, T.; Ohashi, W.; Yagami, T.; Tanaka, S.; Ueda, K.; Sugiura, Y. Arginine-rich peptides An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J. Biol. Chem. 2001, 276, 5836–5840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutta, C.; Chakraborty, K.; Sinha Roy, R. Engineered nanostructured facial lipopeptide as highly efficient molecular transporter. ACS Appl. Mater. Interfaces 2015, 7, 18397–18405. [Google Scholar] [CrossRef] [PubMed]
- Lättig-Tünnemann, G.; Prinz, M.; Hoffmann, D.; Behlke, J.; Palm-Apergi, C.; Morano, I.; Herce, H.D.; Cardoso, M.C. Backbone rigidity and static presentation of guanidinium groups increases cellular uptake of arginine-rich cell-penetrating peptides. Nat. Commun. 2011, 2, 453. [Google Scholar] [CrossRef] [PubMed]
- Khine, Y.Y.; Callari, M.; Lu, H.; Stenzel, M.H. Direct Correlation Between Zeta Potential and Cellular Uptake of Poly (methacrylic acid) Post-Modified with Guanidinium Functionalities. Macromol. Chem. Phys. 2016, 217, 2302–2309. [Google Scholar] [CrossRef]
- Fröhlich, E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomed. 2012, 7, 5577. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.R.; Huang, Y.-w.; Winiarz, J.G.; Chiang, H.-J.; Lee, H.-J. Intracellular delivery of quantum dots mediated by a histidine-and arginine-rich HR9 cell-penetrating peptide through the direct membrane translocation mechanism. Biomaterials 2011, 32, 3520–3537. [Google Scholar] [CrossRef]
- Tünnemann, G.; Martin, R.M.; Haupt, S.; Patsch, C.; Edenhofer, F.; Cardoso, M.C. Cargo-dependent mode of uptake and bioavailability of TAT-containing proteins and peptides in living cells. FASEB J. 2006, 20, 1775–1784. [Google Scholar] [CrossRef] [Green Version]
- Boussif, O.; Lezoualc’h, F.; Zanta, M.A.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.-P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heyes, J.; Palmer, L.; Bremner, K.; MacLachlan, I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Release 2005, 107, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Bacalum, M.; Janosi, L.; Zorila, F.; Tepes, A.-M.; Ionescu, C.; Bogdan, E.; Hadade, N.; Craciun, L.; Grosu, I.; Turcu, I. Modulating short tryptophan-and arginine-rich peptides activity by substitution with histidine. Biochim. Biophys. Acta BBA-Gen. Subj. 2017, 1861, 1844–1854. [Google Scholar] [CrossRef] [PubMed]
- Konate, K.; Josse, E.; Tasic, M.; Redjatti, K.; Aldrian, G.; Deshayes, S.; Boisguérin, P.; Vivès, E. WRAP-based nanoparticles for siRNA delivery: A SAR study and a comparison with lipid-based transfection reagents. J. Nanobiotechnol. 2021, 19, 236. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S. DLS and zeta potential–what they are and what they are not? J. Control. Release 2016, 235, 337–351. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, J.; Yang, X.; Zhao, X.; Xu, H. Oleanolic acid nanosuspensions: Preparation, in-vitro characterization and enhanced hepatoprotective effect. J. Pharm. Pharmacol. 2005, 57, 259–264. [Google Scholar] [CrossRef]
- Aliabadi, H.M.; Maranchuk, R.; Kucharski, C.; Mahdipoor, P.; Hugh, J.; Uludağ, H. Effective response of doxorubicin-sensitive and-resistant breast cancer cells to combinational siRNA therapy. J. Control. Release 2013, 172, 219–228. [Google Scholar] [CrossRef]
- Couto, J.P.; Daly, L.; Almeida, A.; Knauf, J.A.; Fagin, J.A.; Sobrinho-Simões, M.; Lima, J.; Máximo, V.; Soares, P.; Lyden, D. STAT3 negatively regulates thyroid tumorigenesis. Proc. Natl. Acad. Sci. USA 2012, 109, E2361–E2370. [Google Scholar] [CrossRef] [Green Version]
- Messina, J.L.; Yu, H.; Riker, A.I.; Munster, P.N.; Jove, R.L.; Daud, A.I. Activated stat-3 in melanoma. Cancer Control 2008, 15, 196–201. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Sun, X.; Li, X.-L. Expression and clinical significance of STAT3, P-STAT3, and VEGF-C in small cell lung cancer. Asian Pac. J. Cancer Prev. 2012, 13, 2873–2877. [Google Scholar] [CrossRef] [Green Version]
- Jiang, R.; Jin, Z.; Liu, Z.; Sun, L.; Wang, L.; Li, K. Correlation of activated STAT3 expression with clinicopathologic features in lung adenocarcinoma and squamous cell carcinoma. Mol. Diagn. Ther. 2011, 15, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Men, K.; Pan, C.; Gao, Y.; Li, J.; Lei, S.; Zhu, G.; Li, R.; Wei, Y.; Duan, X. Treatment of colon cancer by degradable rrPPC Nano-conjugates delivered STAT3 siRNA. Int. J. Nanomed. 2020, 15, 9875. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Ruan, W.; Qin, M.; Long, Y.; Wan, T.; Yu, K.; Zhai, Y.; Wu, C.; Xu, Y. Intradermal delivery of STAT3 siRNA to treat melanoma via dissolving microneedles. Sci. Rep. 2018, 8, 1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alshamsan, A.; Haddadi, A.; Hamdy, S.; Samuel, J.; El-Kadi, A.O.; Uludag, H.; Lavasanifar, A. STAT3 silencing in dendritic cells by siRNA polyplexes encapsulated in PLGA nanoparticles for the modulation of anticancer immune response. Mol. Pharm. 2010, 7, 1643–1654. [Google Scholar] [CrossRef]












| Sr. No | Conjugate | Sequence | Chemical Formula | Exact Mass | Found (M/Z) |
|---|---|---|---|---|---|
| 1 | Oleyl-(HR)4 | Oleyl-(HR)4-OH | C66H110N28O10 | 1454.896 | 1455.996 [M + H]+ |
| 2 | Oleyl-R1-(HR)4 | Oleyl-R-(HR)4-OH | C72H122N32O11 | 1610.997 | 1612.230 [M + 2 H]+ |
| 3 | Oleyl-R2-(HR)4 | Oleyl-RR-(HR)4-OH | C78H134N36O12 | 1767.098 | 1767.642 [M]+ |
| 4 | Oleyl-R3-(HR)4 | Oleyl-RRR-(HR)4-OH | C84H146N40O13 | 1923.199 | 1925.019 [M + 2 H]+ |
| 5 | Oleyl-R4-(HR)4 | Oleyl-RRRR-(HR)4-OH | C90H158N44O14 | 2079.300 | 2080.422 [M + H]+ |
| 6 | Oleyl-R5-(HR)4 | Oleyl-RRRRR-(HR)4-OH | C96H171N49O14 | 2234.417 | 2236.281 [M + 2 H]+ |
| Peptide | N/P | siRNA (µM) | Peptide Conc. (µM) | W/W Ratio (Peptide/siRNA) | Molar Ratio (Peptide/siRNA) |
|---|---|---|---|---|---|
| Oleyl-(HR)4 | 40 | 1 | 43.6 | 52.5 | 480 |
| Oleyl-R1-(HR)4 | 40 | 1 | 34.9 | 46.5 | 384 |
| Oleyl-R2-(HR)4 | 40 | 1 | 29.1 | 42.5 | 320 |
| Oleyl-R3-(HR)4 | 40 | 1 | 24.9 | 39.7 | 274 |
| Oleyl-R4-(HR)4 | 40 | 1 | 21.8 | 37.5 | 240 |
| Oleyl-R5-(HR)4 | 40 | 1 | 19.4 | 35.9 | 213 |
| Peptide-siRNA Complex at N/P 40. | Z-Ave (d.nm) | SD | PDI | SD |
|---|---|---|---|---|
| Oleyl-(HR)4 | 120.1 | ±1.3 | 0.213 | ±0.03 |
| Oleyl-R1-(HR)4 | 116.9 | ±2.5 | 0.201 | ±0.06 |
| Oleyl-R2-(HR)4 | 115.6 | ±1.6 | 0.217 | ±0.05 |
| Oleyl-R3-(HR)4 | 117 | ±1.8 | 0.229 | ±0.03 |
| Oleyl-R4-(HR)4 | 115.2 | ±2.3 | 0.237 | ±0.07 |
| Oleyl-R5-(HR)4 | 115.6 | ±2.1 | 0.213 | ±0.04 |
| Compound | N/P Ratio | Final Concentration of Peptide in the Complex (µM) | W/W Ratio (Peptide/siRNA) | Molar Ratio (Peptide/siRNA) |
|---|---|---|---|---|
| Oleyl-R2-(HR)4 | 40 | 16 | 42.51 | 320 |
| Oleyl-R3-(HR)4 | 40 | 13.71 | 39.66 | 274.29 |
| Oleyl-R4-(HR)4 | 40 | 12 | 37.53 | 240 |
| Oleyl-R5-(HR)4 | 40 | 10.67 | 35.87 | 213.33 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sajid, M.I.; Mandal, D.; El-Sayed, N.S.; Lohan, S.; Moreno, J.; Tiwari, R.K. Oleyl Conjugated Histidine-Arginine Cell-Penetrating Peptides as Promising Agents for siRNA Delivery. Pharmaceutics 2022, 14, 881. https://doi.org/10.3390/pharmaceutics14040881
Sajid MI, Mandal D, El-Sayed NS, Lohan S, Moreno J, Tiwari RK. Oleyl Conjugated Histidine-Arginine Cell-Penetrating Peptides as Promising Agents for siRNA Delivery. Pharmaceutics. 2022; 14(4):881. https://doi.org/10.3390/pharmaceutics14040881
Chicago/Turabian StyleSajid, Muhammad Imran, Dindyal Mandal, Naglaa Salem El-Sayed, Sandeep Lohan, Jonathan Moreno, and Rakesh Kumar Tiwari. 2022. "Oleyl Conjugated Histidine-Arginine Cell-Penetrating Peptides as Promising Agents for siRNA Delivery" Pharmaceutics 14, no. 4: 881. https://doi.org/10.3390/pharmaceutics14040881